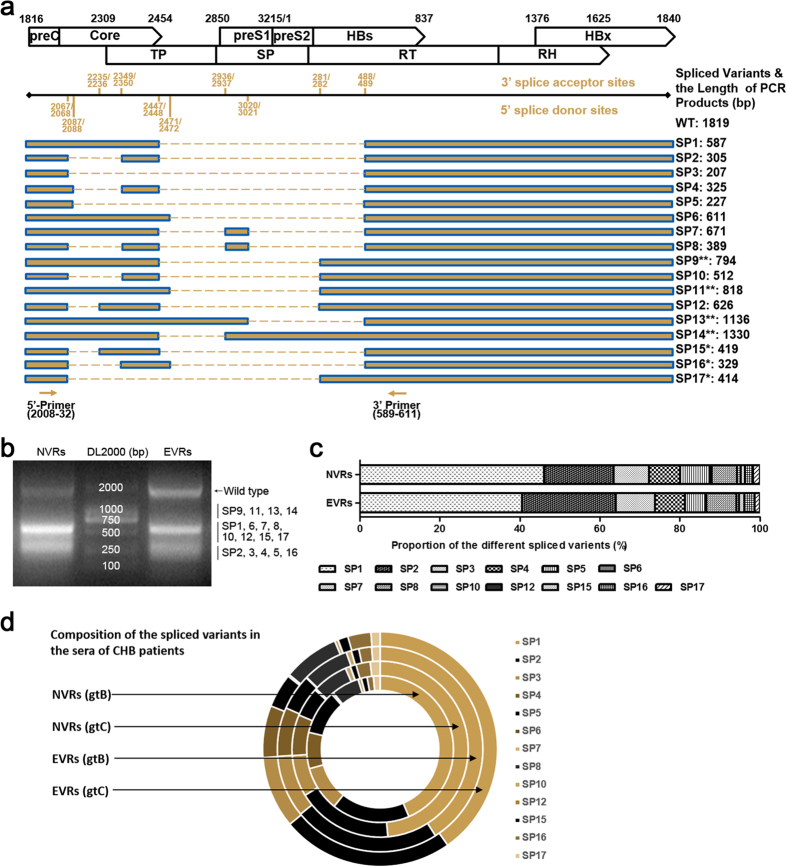
Figure 2. Species of the spHBV variants identified in the sera of CHB patients before IFN therapy.
Schematic representation of the ORFs of HBV genome, the known accept and donor sites for HBV splicing, the reported spHBV variants (**undetected in the current 454 sequencing), the newly identified spHBV variants (*), and the primers designed for PCR amplification of the spHBV variants (a). The HBV DNAs extracted from sera were PCR amplified, and the amplicons were then separately grouped into NVRs and EVRs and pooled, and then subjected to agarose gel electrophoresis (b). The proportions of each of the spHBV variant in NVRs and EVRs according to the 454 sequencing data (c). The composition of spHBV variants in NVRs and EVRs with HBV genotype B and C according to the 454 sequencing data (d).