FIGURE 1.
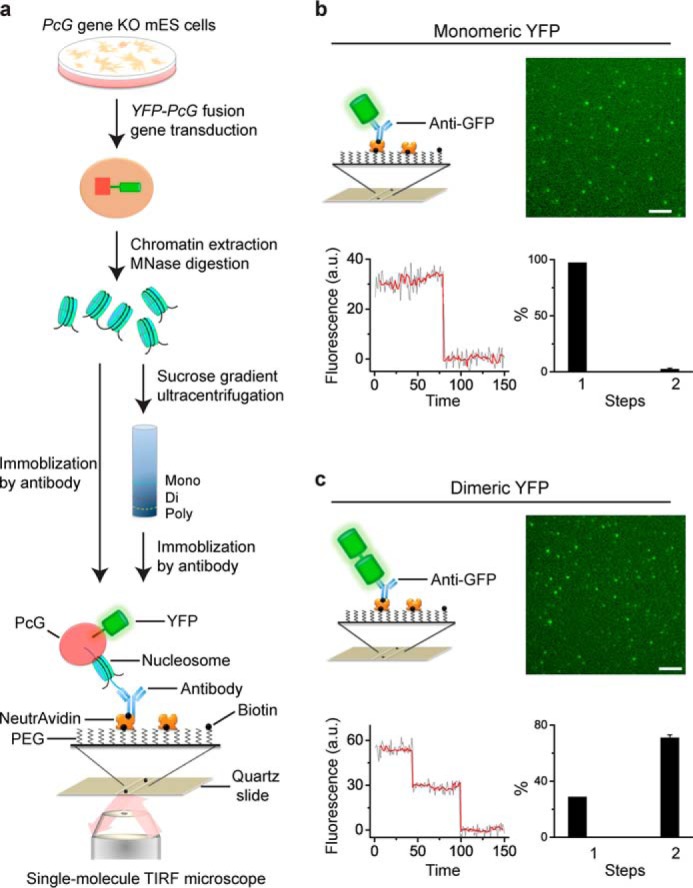
Development of Sm-ChIPi to assess the cellular assembly stoichiometry of PcG complexes on chromatin. a, schematic of the Sm-ChIPi approach. Nucleosomes generated from chromatin are directly immobilized on the surface or are separated by sucrose gradient ultracentrifugation and then immobilized on the surface. b and c, photobleaching behavior of monomeric (b) and dimeric (c) YFPs. The YFP proteins were immobilized by biotinylated anti-GFP antibody via interaction with NeutrAvidin. A sample single image of the acquired sequence is shown. A representative time course of fluorescence emission of the YFP protein is shown (black line). The photobleaching steps were detected by Chung-Kennedy filter (red line). Results are means ± S.D. a.u. denotes arbitrary unit. MNase is micrococcal nuclease. Scale bar, 5 μm.
