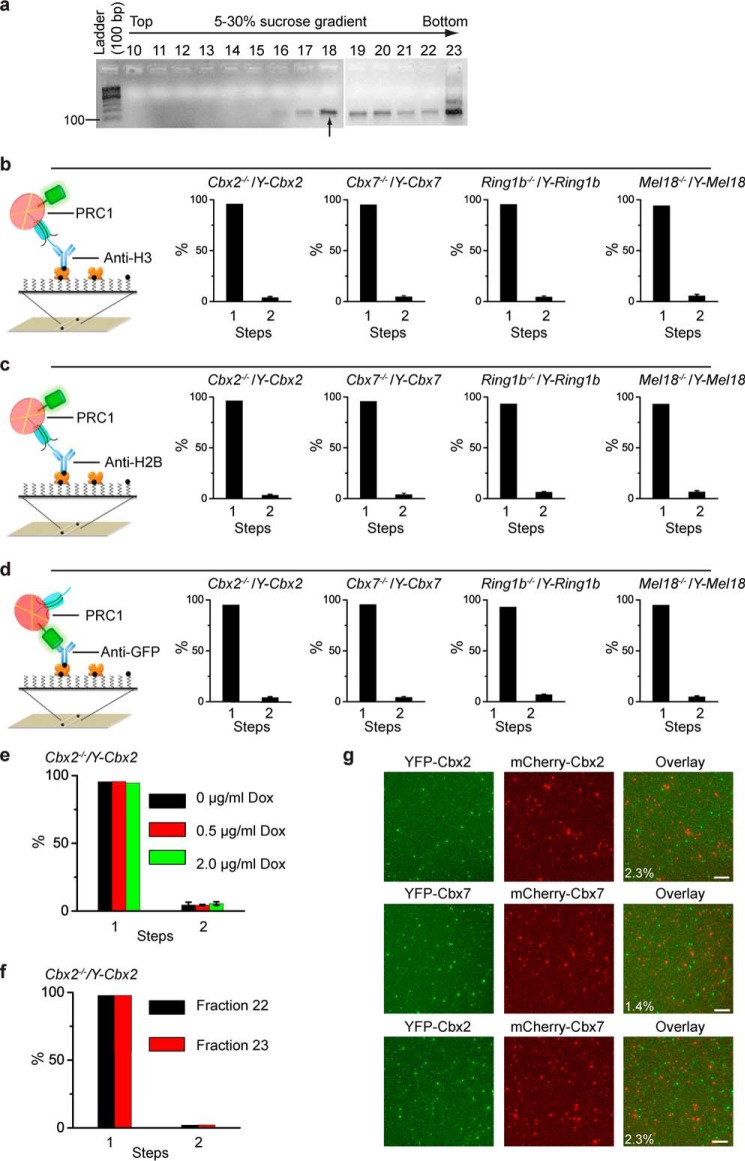
FIGURE 3.
Cellular assembly stoichiometry of YFP-PRC1 proteins on a mononucleosome. a, agarose gel electrophoresis analysis of nucleosomal DNAs extracted from fractions of 5–30% sucrose gradient. Nucleosomes were prepared from Cbx2−/−/Y-Cbx2, Cbx7−/−/Y-Cbx7, Ring1b−/−/Y-Ring1b, and Mel18−/−/Y-Mel18 mES cells. A sample image of agarose gel is shown. Fraction 18 indicated by the arrow below the gel was used for single-molecule TIRF imaging. b–d, percentage of fluorescence photobleaching steps of YFP-Cbx2, YFP-Cbx7, YFP-Ring1b, and YFP-Mel18 on a mononucleosome from fraction 18. The YFP-PRC1·nucleosome complexes were immobilized on the surface by biotinylated antibodies directed against H3 (b), H2B (c), and GFP (d). Results are means ± S.D. e, percentage of fluorescence photobleaching steps of YFP-Cbx2 on a mononucleosome prepared from Cbx2−/−/Y-Cbx2 cells in the presence of Dox concentrations of 0 μg/ml (black bar), 0.5 μg/ml (red bar), or 2.0 μg/ml (green bar). The YFP-Cbx2·nucleosome complexes were immobilized on the surface by biotinylated anti-H3 antibody. Results are means ± S.D. f, percentage of fluorescence photobleaching steps of YFP-Cbx2 on a mononucleosome from fractions 22 and 23. The YFP-Cbx2·nucleosome complexes were immobilized on the surface by biotinylated anti-H3 antibody. Results are means ± S.D. g, single-molecule co-localization analysis. YFP-Cbx2 and mCherry-Cbx2 were stably co-expressed in Cbx2−/− mES cells (top). YFP-Cbx7 and mCherry-Cbx7 were stably co-expressed in Cbx7−/− mES cells (middle). YFP-Cbx2 and mCherry-Cbx7 were stably co-expressed in Cbx7−/− mES cells (bottom). The PRC1·nucleosome complexes from fraction 18 were immobilized by biotinylated anti-H3 antibody. YFP (left) and mCherry (center) were imaged. Overlay of the two images (right) shows 2–3% co-localization. Scale bar, 5 μm.