FIGURE 1.
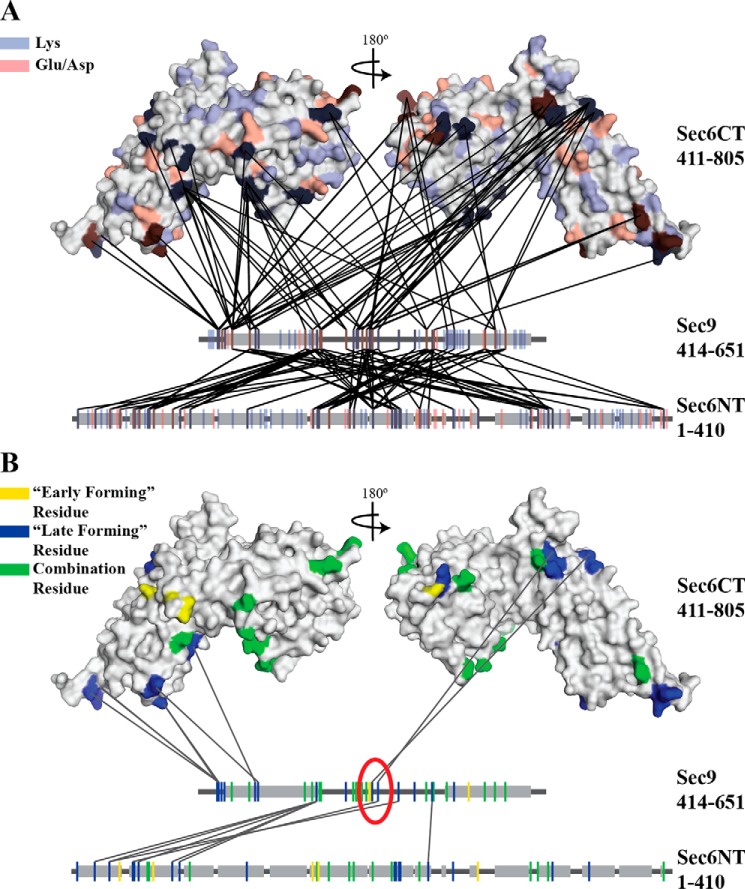
Sec6 and Sec9 cross-link specifically, and different classes of cross-links highlight residues important for the binding interaction. All cross-links identified in each of the experiments described under “Experimental Procedures” were mapped onto the C-terminal structure of Sec6 (amino acids 411–805) (38) and linear segments representing the N-terminal domain of Sec6 and the SNARE domains of Sec9. Gray boxes indicate helical secondary structure (the known SNARE helices on Sec9 and the predicted helical regions of the Sec6 (amino acids 1–410) (SOPM algorithm, Ref. 77)). A, all possible cross-linkable residues (lysine, blue; glutamic acid/aspartic acid, red) are mapped onto the proteins. B, cross-linked residues are colored on the basis of the classes of cross-links identified in Fig. 2A (early-forming, yellow, late-forming, blue, combination residues that participate in both classes, green). Only the late-forming cross-links are shown. On the basis of the hypothesized model presented in Fig. 2B, residues likely to be involved in the binding interaction are circled in red. The sec9-142 mutation is located between the two blue residues (Lys-533 and Glu-537) in the red circle.