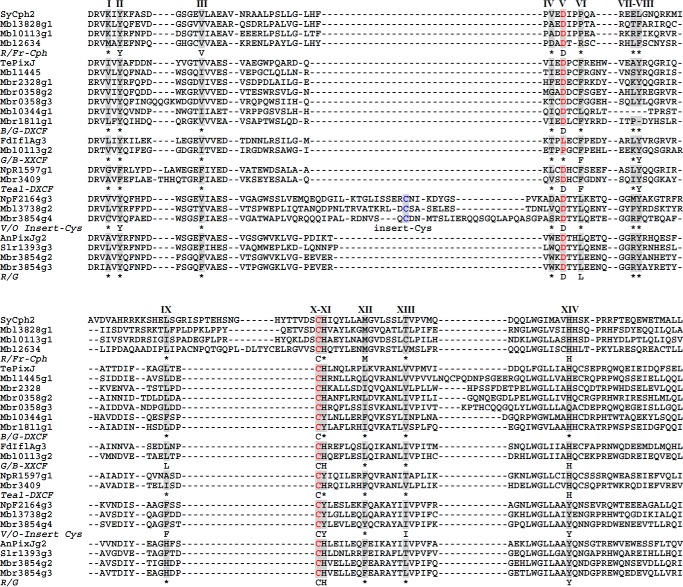
FIGURE 4.
Multiple protein sequence alignment of bilin-binding GAFs from Microcoleus B353. CBCR GAF subclasses are clustered based on spectral properties of the nearest known relative. R/FR, red/far-red GAFs include Cph1 and knotless Cph2 phytochromes; B/G-DXCF, blue/green GAFs originally identified with 2nd Cys in Asp motif (DXCF) also includes blue/orange (Mbr0358g2), violet/orange (Mbr0358g3), violet/yellow (Mbr2328), and blue/blue (Mbl0344g1) CBCRs; teal-DXCF, GAFs with PVB chromophore contain teal absorbing photostate converted from green (green/teal, NpR5113g1) or blue (blue/teal, NpR1597g1, Tlr1999g1) light absorbing forms; G/B-XXCF, this subfamily consists of green/blue GAFs with Phe (Mbl0113g2, NpR5313g2) or Leu (FdIflAg3) variation in the Asp motif; V/O-Insert Cys, violet/orange GAFs with 2nd Cys residue located at the insert region include violet/orange (NpF2164g3), near-UV/blue (NpF2164g2 and NpR1597g2), and near-UV/green (Mbr3854g4 and Mbl3738g2) photocycles; R/G, red/green GAFs such as AnPixJg2 and Slr1393g3. Amino acid residues contacting (or near) the chromophore based on the three-dimensional structure of AnPixJ2 are marked with Roman numerals. Consensus residues are obtained from multiple sequence alignments from experimentally characterized CBCR GAFs. *, variable residues.