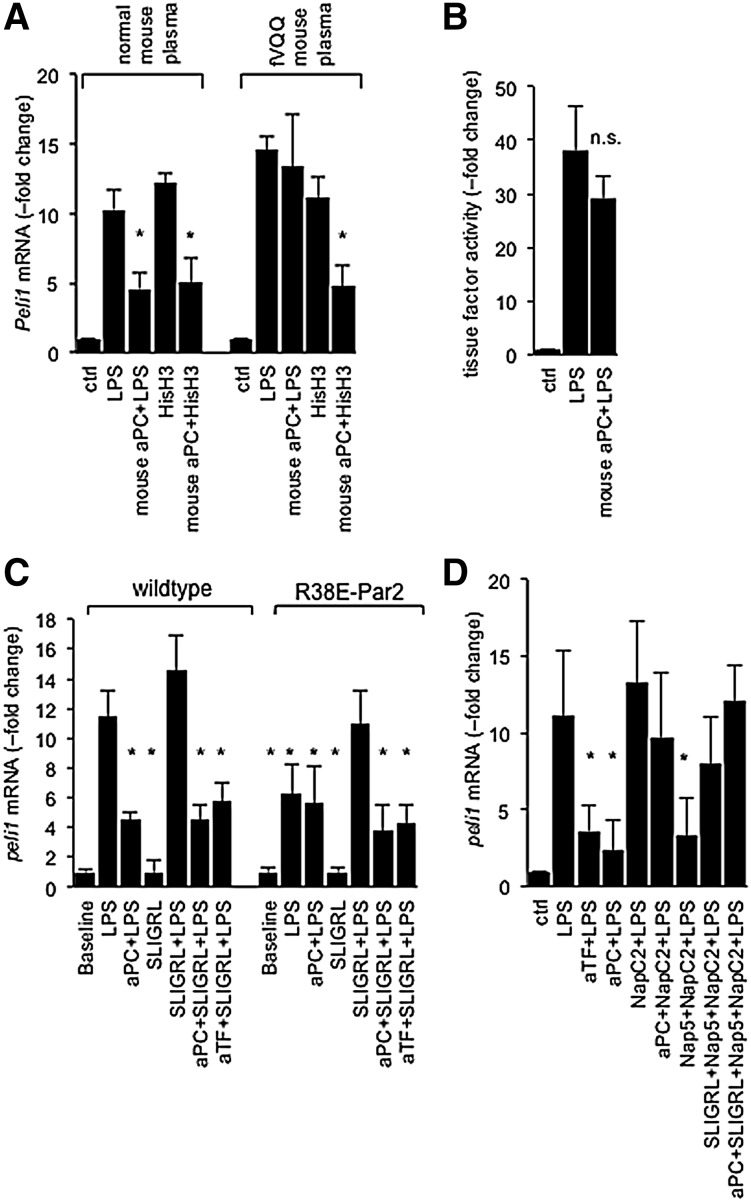
Figure 4.
aPC inhibits PAR2 activation by the ternary TF-fVIIa-fXa complex. (A) aPC suppresses histone-induced Peli1 expression independent of fV. Quantitative PCR measurement of Peli1 mRNA in RAW cells treated for 3 hours with the indicated reagents (100 ng/mL of LPS; LPS plus 100 ng/mL of mouse 5A-aPC [mouse aPC+LPS]; 30 μg/mL of calf thymus histone H3 [HisH3]; 30 μg/mL of HisH3 plus 100 ng/mL of mouse 5A-aPC [mouse aPC+HisH3]). *P = .05 by Student t test, compared to cells treated with LPS alone. (B) TF activity in was measured in RAW cells via the rate of fXa generation in a 2-stage amidolytic assay for fXa activity at baseline and after 3 hours of exposure to LPS or LPS plus 5A-aPC (mouse aPC+LPS) in the presence of normal plasma. At this time, cells were washed and incubated with recombinant mouse fVII and fX. Data are corrected for background activity (no fVII) and represent the average ± standard deviation of 6 independent experiments, with triplicate measurement per sample. n.s., not significant. (C) Peli1 mRNA regulation by LPS in BMDCs prepared from wild-type mice or a knockin mouse strain expressing the cleavage-resistant R38E-PAR2 variant. Data represent the average ± standard deviation from 3 independent experiments. *P < .05 by Student 2-tailed t test, as compared to LPS-treated wild-type cells. (D) RAW cells were incubated for 3 hours with LPS (100 ng/mL) and the indicated reagents (anti-murine TF antibody [aTF], 5 μg/mL; mouse 5A-aPC [aPC], 100 ng/mL; Nap5, 500 nM; NapC2, 500 nM; and SLIGRL, 20 μM), and Peli1 mRNA was quantitative by real-time PCR analysis. Data represent the average ± standard deviation from 6 independent experiments. *P < .05 by Student t test, compared to cells treated with LPS alone. PCR, polymerase chain reaction.