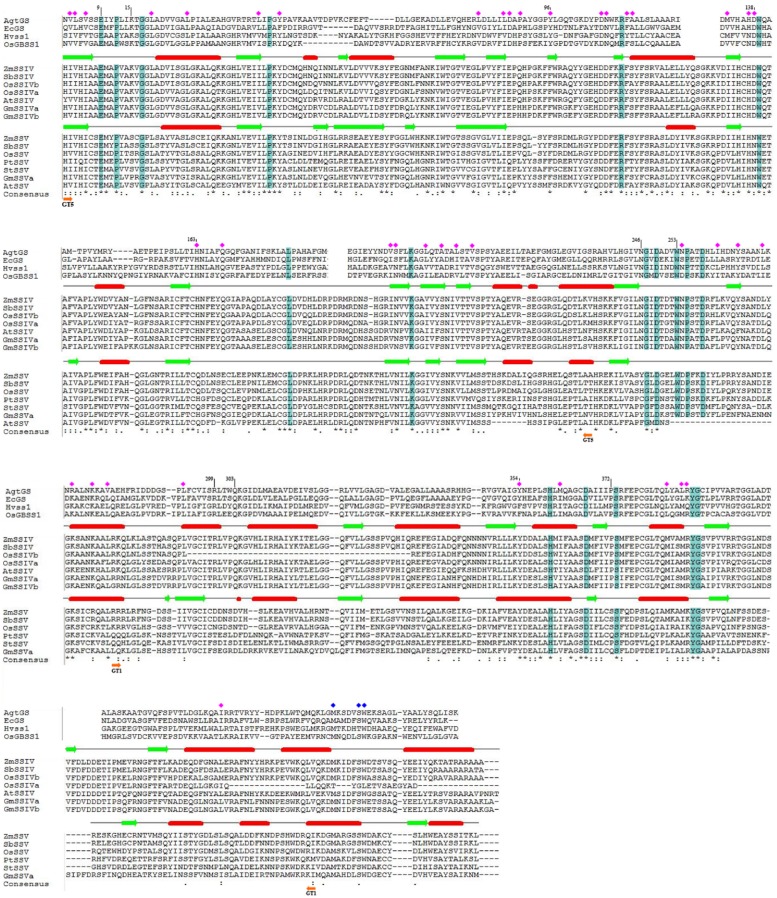
FIGURE 4.
The protein sequence alignment of SSIV and SSV in eudicots and monocots, for C-terminus conserved region. For further investigation the important functional sites, partial alignment with GSs and SSs of which the 3D structure were determined were showed. Moreover, the secondary structure of ZmSSIV and ZmSSV were predicted, α-helical indicated by red cylinder, and β-strand indicated by green arrows. The identical amino acid residues in a colum were highlighted in blue, or noted by blue squares(except the differing residues of OsSSIVa end), and conserved amino acid residues were noted by pink squares. In the line of the consensus, identical and conserved residues between SSIV and SSV were marked with asterisks (∗) and dots/colons (./:), respectively, while, dashes indicate no residue. The boundary regions of GT5 and GT1 were marked with orange arrows.