Fig. 1.
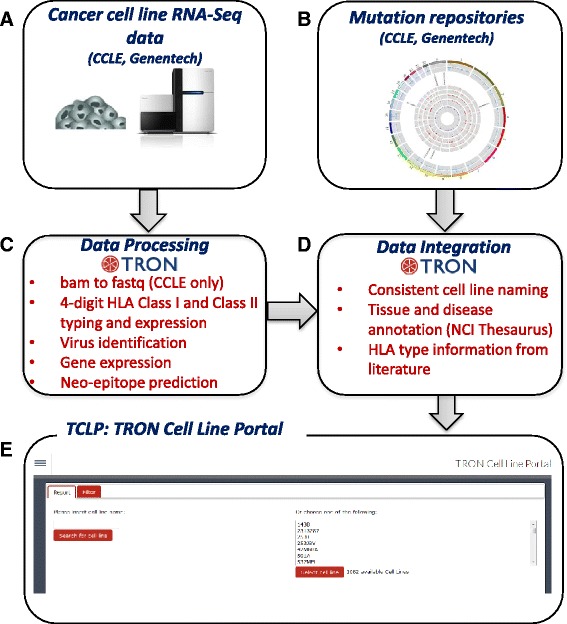
Data integration and computational workflow. RNA-Seq data from 1,083 human cancer cell lines is downloaded from CCLE and Genentech (a) and mutation information for the cell lines is retrieved (b). The RNA-Seq reads are processed by our in-house pipeline (c), consisting of HLA typing and quantification, virus identification, gene expression analysis, and neo-epitope prediction. These data are integrated using consistent cell line names as primary identifier and annotate tissue and disease information using the onotology NCI Thesaurus (d). The results are freely accessible in the TRON Cell Line Portal (e) at http://celllines.tron-mainz.de
