Abstract
Fragile X-associated disorders are Repeat Expansion Diseases that result from expansion of a CGG/CCG-repeat in the FMR1 gene. Contractions of the repeat tract also occur, albeit at lower frequency. However, these contractions can potentially modulate disease symptoms or generate an allele with repeat numbers in the normal range. Little is known about the expansion mechanism and even less about contractions. We have previously demonstrated that the mismatch repair (MMR) protein MSH2 is required for expansions in a mouse model of these disorders. Here, we show that MSH3, the MSH2-binding partner in the MutSβ complex, is required for 98% of germ line expansions and all somatic expansions in this model. In addition, we provide evidence for two different contraction mechanisms that operate in the mouse model, a MutSβ-independent one that generates small contractions and a MutSβ-dependent one that generates larger ones. We also show that MutSβ complexes formed with the repeats have altered kinetics of ATP hydrolysis relative to complexes with bona fide MMR substrates and that MutSβ increases the stability of the CCG-hairpins at physiological temperatures. These data may have important implications for our understanding of the mechanism(s) of repeat instability and for the role of MMR proteins in this process.
Introduction
The Fragile X (FX)-related disorders (FXDs) are Repeat Expansion Diseases that result from expansion of a microsatellite with the repeat unit CGG/CCG. The microsatellite is located in the 5′ UTR of the FMR1 gene [reviewed in (1)]. Alleles with 55–200 repeats are referred to as premutation (PM) alleles and such alleles are prone to expansion or contraction in humans and in an FXD mouse model (2–4). Moderate expansions can result in larger PM alleles that modulate the risk of Fragile X-associated tremor/ataxia syndrome and Fragile X-associated primary ovarian insufficiency, two clinical conditions that affect PM carriers. More extensive expansions into the full mutation range (>200 repeats) result in Fragile X syndrome, a form of intellectual disability. Contractions are also clinically relevant because they can result in tissue mosaicism (5–10) that can complicate risk assessment. Contractions can also modulate disease risk or even result in reversion to a normal allele (11–15). The molecular basis of these expansions and contractions is not known. We and others have shown that the individual strands of the FX repeat form hairpins and other folded structures that are similar to those formed by other disease-associated repeats (16–26). These structures are thought to be the substrates upon which the expansion and contraction pathways act. Interruptions to the purity of the CGG/CCG-repeat tract in the form of one or more interspersed AGG/CCT triplets are seen in some FMR1 alleles. These interruptions would be predicted to reduce hairpin stability and have been shown to reduce risk of expansion in humans (27–29).
Two basic types of expansion models have been proposed, the first in which instability results from difficulties with replication of the repeat-containing region during normal chromosome duplication and a second that invokes some form of aberrant DNA repair (30). However, an effect of maternal age on the risk of expansion in humans is seen, suggesting that expansion occurs in oocytes (28), a cell in which DNA replication does not occur. Thus, a problem with DNA repair rather than chromosomal replication may be responsible for expansion, at least in some cells. However, we have also shown that in mice somatic expansion only occurs when the PM allele is on the active X chromosome (31) and a re-examination of human data (32) suggests that the same is true in women. Thus, transcription or transcriptionally competent chromatin plays an important role in the expansion process (31).
We have previously shown that this transcription dependence is not related to a role for Transcription Coupled Repair (4). Rather, we have shown that the mismatch repair (MMR) protein MSH2 is required for all germ line and somatic expansions in the mouse model (33). A similar requirement for MSH2 has been observed for expansion in other mouse models [reviewed in (34)]. However, whether MSH2 acts to promote expansions in the FXD mouse via its interaction with MSH3 in the MutSβ complex, one of the two MSH2-containing complexes in mammalian cells, or via its interaction with MSH6 in the MutSα complex, the second MSH2-containing complex, was not known. Furthermore, very little is known about the mechanism responsible for contractions. In fact, to date no proteins or pathways have been identified that are responsible for the generation of contractions in humans or in mouse models.
We show here that MSH3, and thus MutSβ, is required for 98% of germ line expansions and all somatic expansions in a mouse model for the FXDs. In addition to a role of MutSβ in generating expansions, we also uncovered two additional unappreciated aspects of CGG-repeat instability in these animals. First, we show that in mice wild type (WT) for Msh3, two discrete contraction size classes can be seen, one involving the loss of ∼1–2 repeats and a second that results in the loss of larger repeat numbers. Second, we demonstrate a hitherto undescribed role for MutSβ in repeat instability in mice, namely its ability to promote the formation of larger contractions. The fact that MutSβ promotes the generation of one class of contractions and not the other, along with our biochemical studies on binding of MutSβ to the FX repeats, has interesting implications for models of the mechanism of repeat instability.
Results
Loss of MSH3 eliminates all somatic expansions
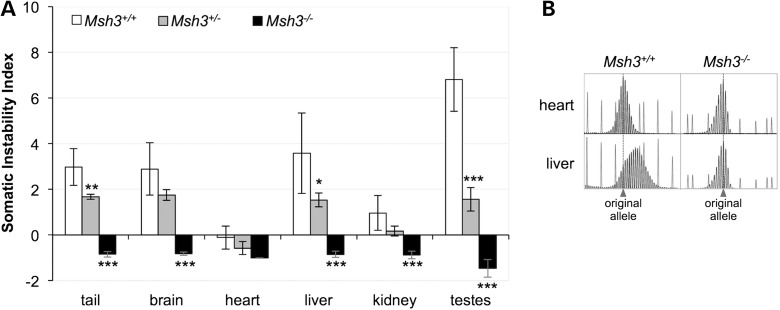
We examined the effect of the loss of MSH3 on the extent of expansion in the somatic tissue of FXD male mice with ∼140 CGG/CCG repeats. We used the somatic instability index (SII) as a quantitative measure of the extent of repeat expansion (35). In Msh3+/+ animals, the heart, an organ that shows very little somatic instability, has an SII of −0.1 (Fig. 1). The negative value reflects the contribution to the SII of PCR products that are smaller than the original allele. These products are present at the same levels in the PCR profiles of all organs tested and in animals of all ages (3,4,36) and thus likely represent strand-slippage products produced by PCR across the repeats as described previously for the FX repeats (33) as well as many other short tandem repeats (37–39). In organs other than the heart, the SII ranged from 1.0 (kidney) to 6.8 (testes). As can be seen in Figure 1B, most of the PCR products larger than the original allele seen in the heart of an Msh3+/+ mouse are absent from the PCR profile generated from DNA purified from the heart of an Msh3−/− mouse. However, the PCR products smaller than the original allele were indistinguishable from those seen in Msh3+/+ mice. Almost all of the PCR products larger than the original allele are also lost from other, more expansion-prone organs. For example, the PCR profile for liver was indistinguishable from that of heart in Msh3−/− animals (Fig. 1B). In fact, all organs of Msh3−/− mice showed a slight negative SII similar to what was seen in heart (Fig. 1A). Thus, expansions were essentially eliminated in Msh3−/− animals and because no novel PCR products smaller than the original allele were observed in any organ, the loss of expanded alleles cannot be explained by an increase in contractions. Thus, MSH3, and therefore MutSβ, is required for the generation of somatic expansions. Even in Msh3+/− animals, the SII was significantly lower in tail, liver and testes than that in Msh3+/+ animals indicating that the loss of even one Msh3 allele is sufficient to reduce the expansion frequency (Fig. 1A).
Figure 1.
Loss of MSH3 reduces somatic expansions. (A) The SII for the organs of six 6-month-old Msh3+/+, Msh3+/− and Msh3−/− males was determined and the average SII for each organ plotted. The initial repeat number in these mice was ∼140. The negative value for the heart in animals of each genotype and for the remaining organs in Msh3−/− mice reflects the strand-slippage or stutter products that are typically seen when large repeat tracts are amplified rather than contraction events. The SIIs for Msh3−/− animals that are significantly different from WT animals are marked with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001). (B) Comparison of the GeneMapper PCR profiles for the heart and liver of one Msh3+/+ and one Msh3−/− mouse.
Loss of MSH3 eliminates almost all germ-line expansions
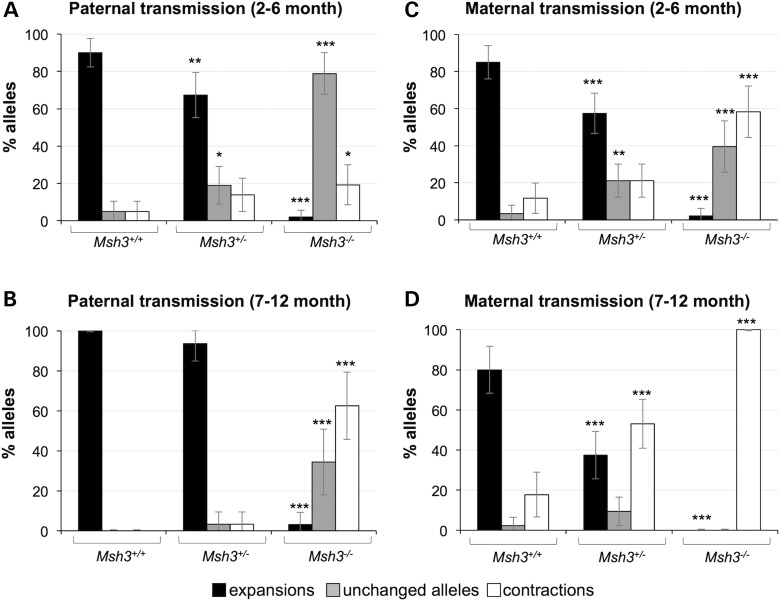
In Msh3+/+ males, the expansion pathway predominates as evidenced by the fact that 90% of alleles transmitted by 2- to 6-month-old Msh3+/+ males had expansions (Fig. 2A). In contrast, in the progeny of Msh3−/− fathers of the same age, only 2% did. This decrease in expansions in the progeny of Msh3−/− fathers is associated with an increase in the frequency of unchanged alleles [79% up from 5% seen in the progeny Msh3+/+ fathers (P = 0.0001); Fig. 2A]. Thus, our data would be consistent with a role of MSH3, and thus MutSβ, in promoting expansions. The fact that unchanged alleles predominate when expansions are lost in Msh3−/− mice confirms our earlier hypothesis that the contraction pathway is not very active in young males (4). The proportion of contractions is significantly increased in the progeny of 7- to 12-month-old Msh3−/− males (Fig. 2B), consistent with the idea that the contraction pathway becomes more prominent with age in these animals (4).
Figure 2.
Loss of MSH3 reduces paternally and maternally transmitted intergenerational expansions. The number of expansions, contractions and unchanged alleles seen on transmission of a PM allele with ∼140 repeats from WT, Msh3+/− and Msh3−/− parents was determined in the progeny of 2- to 6-month-old (A) and 7- to 12-month-old (B) males and in the progeny of 2- to 6-month-old (C) and 7- to 12-month-old (D) females. The error bars represent the 95% confidence intervals. The allele classes that are significantly different from WT animals are marked with asterisks (*P < 0.05; **P < 0.01; ***P < 0.001).
The proportion of contracted and unchanged alleles in the progeny of 2- to 6-month-old Msh3−/− mice also contrasts with what was seen in the progeny of age-matched Msh2−/− animals where both unchanged and contracted alleles increased to similar extents (33). As Msh3−/− mice lack MutSβ, but still produce MutSα, whereas Msh2−/− mice lack both MutSβ and MutSα, the difference in the distribution of the residual alleles in Msh2−/− mice compared with Msh3−/− mice likely reflects the effect of the loss of protection by MutSα against repeat contractions. Thus, MutSα may be a key player in the error-free repair pathway that we had previously identified (36). This hypothesis is currently being tested in mice lacking MSH6.
Loss of MSH3 in 2- to 6-month-old females also resulted in a significant reduction in expansions (Fig. 2C). However, in contrast to the relatively low level of contractions seen in the offspring of males, 58% of the residual alleles were smaller than the parental allele. Furthermore, the contraction frequency increased with age such that in the progeny of 7- to 12-month-old Msh3−/− females, only contractions were seen (Fig. 2D). This is consistent with our previous observations that the contraction pathway is more active in the female germ line than it is in the male germ line even in animals WT for the MMR proteins (4). The contraction pathway is thus able to compete more effectively with the error-free pathway for the instability substrate when the expansion pathway is eliminated.
Msh3+/+ mice show a bimodal distribution of germ line contraction sizes
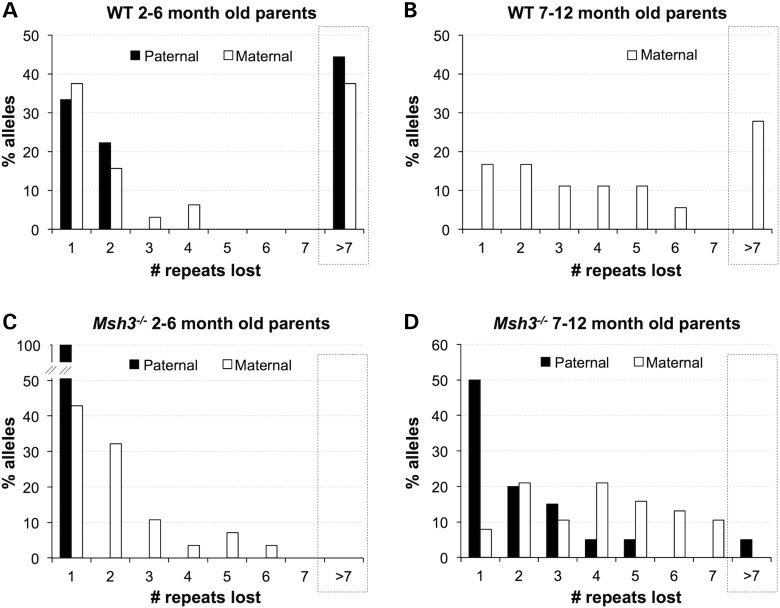
Visual inspection of the contractions seen in the progeny of 2- to 6-month-old Msh3+/+ parents suggested a bimodal distribution of contraction sizes. This was confirmed using Hartigans’ dip test (40) that indicated a significant deviation from unimodality consistent with a bimodal distribution, for both paternal (P = 0.029) and maternal (P = 0.041) transmission. Based on the minima seen in the associated density plots (−7 for females and −10 for males), the alleles were divided into 2 groups, 1 involving repeats having lost <7 repeats and 1 involving the loss of >7 repeats (Fig. 3A). The first group had an average loss of ∼1–2 repeats, whereas the second group had an average loss of 12 repeats for females and 17 for males. In the case of paternal transmissions, 56% of contractions fell into the first group, whereas 44% of contractions fell into the second group (black bars in Fig. 3A). In the case of maternal transmission, 53% of contractions fell into the first group, whereas 37% of contractions fell into the second group (white bars in Fig. 3A). This bimodal distribution was not noted previously because the low frequency of contractions in WT animals makes the accumulation of statistically significant sample numbers difficult.
Figure 3.
The distribution of contraction sizes in the progeny of Msh3+/+ and Msh3−/− males and females. The distribution of the numbers of repeats lost was plotted for the contracted alleles in the progeny of 2- to 6-month-old Msh3+/+ males (n = 9) and females (n = 32) (A), 7- to 12-month-old Msh3+/+ females (n = 18) (B), 2- to 6-month-old Msh3−/− males (n = 10) and females (n = 28) (C) and 7- to 12-month-old Msh3−/− males (n = 20) and females (n = 38) (D). No contractions were seen in 7- to 12-month-old Msh3+/+ males.
As previously reported (4,33), no contractions are seen in the progeny of older Msh3+/+ males (Fig. 2B). This is due to the high frequency of expansion that persists throughout life in male mice (4). The net result is that, in the progeny of older males, even alleles that once had sustained contractions have now undergone one or more rounds of expansion (4). Msh3+/+ females in this age group still show contractions, perhaps because the contraction frequency increases in prominence in females with age (4). However, the progeny of Msh3+/+ females show a broader distribution of contraction sizes with less clear evidence for bimodality than do the progeny of 2- to 6-month-old females (Fig. 3B). This may be because in older mothers each transmitted allele has been subjected to multiple rounds of expansion and/or contraction that complicate the data interpretation.
The bimodal distribution of contractions that is apparent in younger animals may indicate the operation of two different contraction mechanisms, one that gives rise to small contractions and one that is responsible for larger ones.
Loss of MSH3 increases the contraction frequency but reduces the average contraction size
While the number of contractions transmitted was ∼3-fold higher in 2- to 6-month Msh3−/− males than it was in similarly aged WT males, careful analysis demonstrated that all of these contractions involved the loss of a single repeat unit (Fig. 3C; Fisher's exact test for the difference between small (<7) and large (>7) contractions in Msh3+/+ and Msh3−/− males P = 0.0325). Similarly, while 2- to 6-month-old Msh3−/− females transmitted ∼5-fold more contractions than 2- to 6-month-old Msh3+/+ females, no large contractions were seen (Fig. 3C; Fisher's exact test for the difference been small and large contractions in Msh3+/+ and Msh3−/− females, P = 0.0015). However, the transmitted contractions were slightly bigger on average than the contractions transmitted by Msh3−/− males (−2.11 versus −1.00; t-test: P = 0.0178). This could be due to the higher contraction frequency seen on maternal transmission that increases the likelihood that some alleles undergo more than one round of contraction (compare Fig. 2A and C).
As no contracted alleles are seen in the progeny of older Msh3+/+ males, we were unable to compare the contraction sizes in Msh3+/+ and Msh3−/− males in the 7- to 12-month-old range. However, as can be seen from Figure 3D, the progeny of 7- to 12-month-old Msh3−/− males had a distribution of contraction sizes that was still displaced toward smaller alleles than the progeny of 2- to 6-month-old Msh3+/+ males with only 5% of alleles having >7 repeats. As can be seen in Figure 3D, in the progeny of 7- to 12-month-old Msh3−/− females, the average contraction size was also smaller than that in Msh3+/+ mice of the same age with no large contractions being found (Fisher's exact test for the difference been small and large contractions, P = 0.0001). Taken together our data thus show that the loss of Msh3 leads to preferential loss of larger contractions but does not eliminate contractions altogether.
MutSβ binds to and stabilizes the secondary structures formed by the FX repeats
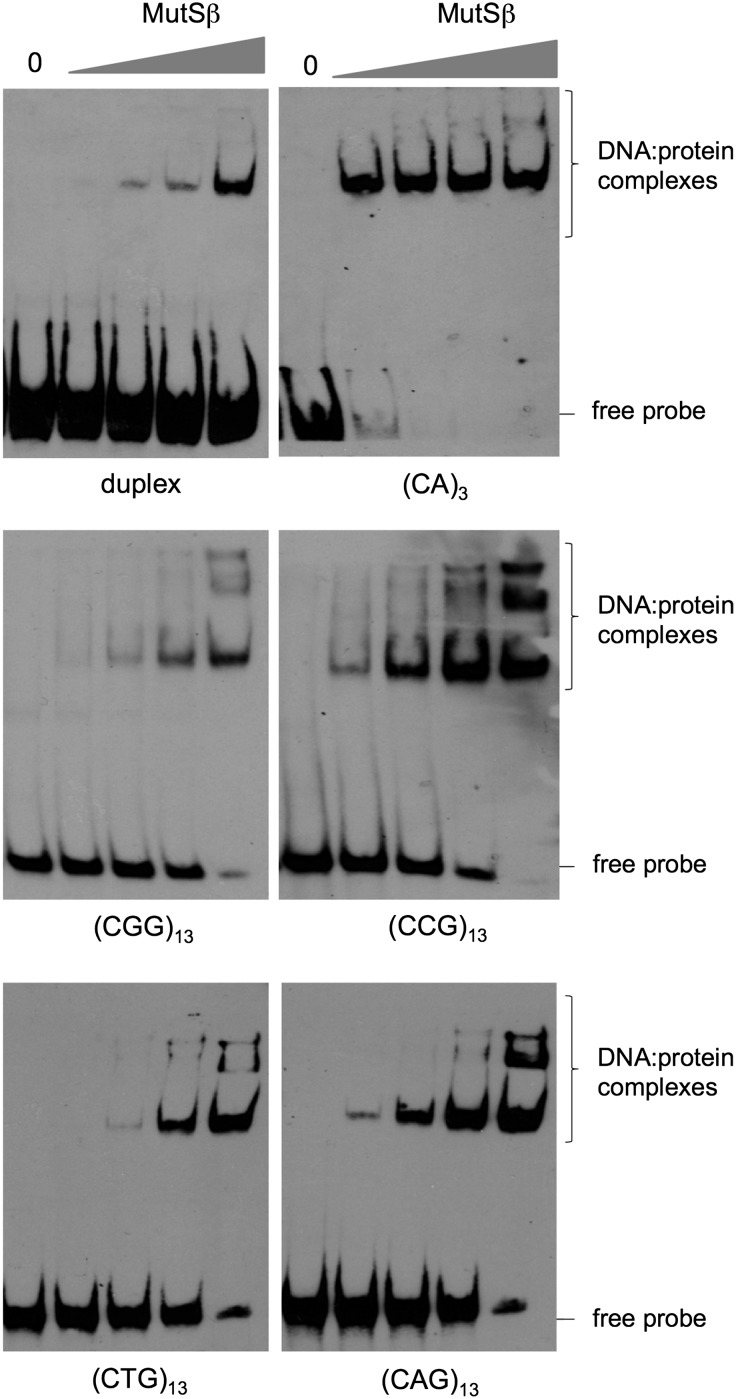
As promoting expansions and contractions is inconsistent with the canonical role of MMR in protecting against microsatellite instability, we initiated biochemical studies aimed at understanding other ways in which MutSβ might be acting. We first generated substrates containing a loop-out of (CCG)13 or (CGG)13 that were modeled on those used previously to examine MutSβ binding to CAG-repeats (41) and shown in Table 1. We included substrates with (CAG)13 or (CTG)13-repeat loop-outs for comparison. We then tested the ability of purified MutSβ to bind to these structures. As can be seen in Figure 4, MutSβ binds as well to CGG- and CCG-repeats as it does to CAG- and CTG-repeats. In fact, if anything, the binding of CCG-repeats is more extensive than binding to the other repeats as evidenced by the binding seen at lower concentrations of MutSβ as well as by the ratio of bound to unbound probe. Of each pair of loop-outs, the loop-out forming the hairpin with the most unstable mismatch was the one that was bound more strongly (i.e. CCG>CGG; CAG>CTG). Evidence of multiple DNA–protein complexes could be seen on each DNA substrate, consistent with the idea that more than one MutSβ dimer is able to bind such loop-outs as previously reported for CAG-loop-outs (41).
Table 1.
Oligonucleotides used in this study
| Name | Sequence | Assay |
|---|---|---|
| DuplexBSa | 5′ CTGCCTCAAGTGTTCGGACTCTGCCTCAAATGA CGGTAGTCAACGGCTTGGACGGTAGT 3′ |
EMSA/ATPase |
| DuplexTS | 5′ ACTACCGTCCAAGCACGTTGACTACCGTCATTT GAGGCAGAGTCCGAACACTTGAGGCAG 3′ |
EMSA/ATPase |
| (CNG)13TSb | 5′ ACTACCGTCCAAGCACGTTGACTACCGTCACNG CNGCNGCNGCNGCNGCNGCNGCNGCNGCNGCNG CNGTTTGAGGCAGAGTCCGAACACTTGAGGCAG 3′ |
EMSA/ATPase |
| (CA)3TS | 5′ ACTACCGTCCAAGCACGTTGACTACCGTCACAC ACATTTGAGGCAGAGTCCGAACACTTGAGGCAG 3′ |
EMSA/ATPase |
| (CCG)10 | 5′ ROX-CCGCCGCCGCCGCCGCCGCCGCCGCCGC CG-IOWA Black RQ 3′ |
Thermal melting |
aThis oligonucleotide was labeled at the 5′ end with biotin during synthesis for use in EMSA reactions.
bN represents either A, G, C or T.
Figure 4.
MutSβ binds to the individual strands of the FX repeat. Purified human MutSβ was added to reaction mixtures containing either a fully heteroduplex molecule (duplex) or otherwise duplex oligonucleotides containing the indicated repeat loop-outs at concentrations of 200 pM, 400 pM, 1 and 4 nm as described in the Materials and Methods. The DNA and DNA–MutSβ complexes were then analyzed as described in the Materials and Methods. Note that while some MutSβ binding to duplex DNA, a poor MMR substrate, can be seen (upper left panel), this binding is relatively inefficient as evidenced by the large fraction of unbound (free) probe.
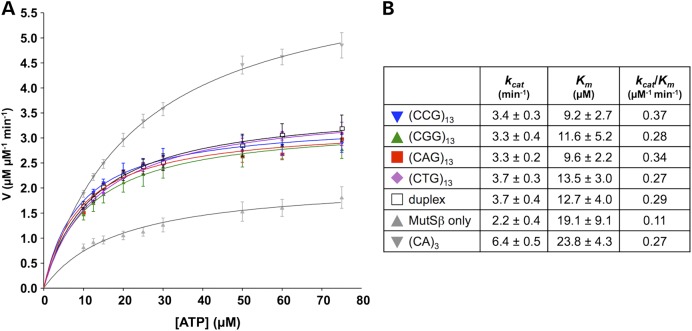
Recent works suggest that ATP binding and hydrolysis by MutSβ are differentially modified by the substrates of different repair pathways (42–44). Specifically, it has been suggested that substrates of different repair pathways induce specific conformational changes in the DNA-binding domains of MutSβ that are then relayed to the ATPase domains resulting in changes in the kinetics of ATP hydrolysis (42). As can be seen in Figure 5 and consistent with what was reported for a CAG-loop-out (41), binding to either a CGG-loop-out or a CCG-loop-out resulted in altered kinetics of ATP hydrolysis relative to binding to a (CA)3 loop-out that is a bona fide MMR substrate (45). Thus, differences likely exist between the conformation of MutSβ when bound to the FX loop-outs and the conformation of MutSβ bound to a bona fide MMR substrate that affects ATP hydrolysis. This altered MutSβ conformation may result in less efficient signaling to proteins downstream in the MMR pathway or in more efficient signaling to an alternate repair pathway.
Figure 5.
Binding of CGG- and CCG-repeats affects the kinetics of ATP hydrolysis by MutSβ. Steady-state ATP hydrolysis was measured using a TLC-based assay as described in the Materials and Methods section. MutSβ without any added DNA was used as a negative control. A oligonucleotide containing a (CA)3 loop-out that elicits efficient ATP hydrolysis by MutSβ (45) was used as a positive control. Duplex is a fully Watson-Crick base-paired oligonucleotide, i.e. containing no mismatches. Its sequence corresponds to the duplex regions of the repeat-containing oligonucleotides (including (CA)3) and serves as a control for MutSβ binding to the free ends of the duplex regions.
To assess the effect of MutSβ binding on the stability of the FX-repeat structures, we monitored the thermal denaturation of the oligonucleotide in the presence of BSA or MutSβ. As the hairpin-to-single-stranded transition of even a very short CGG-repeat oligonucleotide occurs at temperatures above the denaturation temperature of the most proteins (16,46), we limited our study to the CCG-repeat. The 5′ end of a (CCG)10 oligonucleotide was labeled with 5-carboxy-X-rhodamine (ROX™), a fluorescence donor and the 3′ end was labeled with IOWA Black® RQ, a fluorescence acceptor/quencher. This enabled the stability of the hairpins to be assessed in the presence of MutSβ by monitoring the increase in the fluorescence signal at the ROX™ emission wavelength with increasing temperature. The oligonucleotide was denatured and cooled under conditions in which the repeats are known to form hairpins (17–21,47,48). The oligonucleotide was then mixed with MutSβ and subjected to increasing temperatures as described in the Materials and Methods. Increasing temperatures resulted in a progressive increase in fluorescence at 608 nm consistent with melting of the secondary structure formed by the CCG-repeat. The melting curves obtained for both protein-CCG-repeat mixtures fit a two-state model (Supplementary Material, Fig. S2). The thermodynamic parameters derived from analysis of the melting curves are shown in Table 2. As can be seen from this table, the presence of MutSβ resulted in higher ΔG at 37°C than is seen in the presence of BSA suggesting that MutSβ increases the stability of the CCG-repeat structure at physiological temperatures.
Table 2.
Comparison of the free energy (ΔG) at 37°C and enthalpy (ΔH) required to denature the secondary structure formed by CCG-repeats
| Protein | ΔG (37°C) kcal/mol | ΔHm kcal/mol |
|---|---|---|
| BSA | 1.88 ± 0.08 | 28.4 ± 0.9 |
| MutSβ | 3.48 ± 0.18 | 52.4 ± 4.1 |
Discussion
We have previously shown that MSH2 is required for all paternal and maternal germ line expansions as well as for somatic expansions. We show here that loss of MSH3 eliminates 98% of germ line and all somatic repeat expansions in these animals (Figs 1 and 2). As MSH3 acts together with MSH2 in the MutSβ complex, our data demonstrate that MutSβ is required for almost all expansions in the FXD mouse model. This would be consistent with what has been found in some mouse models of other Repeat Expansion Diseases (49) but not others (50,51). As MSH2 does not act independently of MSH3 and MSH6 in eukaryotes, it seems most likely that the remaining MSH2-dependent MSH3-independent germ line expansions involve MSH6 and thus MutSα. MutSα has been implicated in somatic expansion in a mouse model of the GAA/TTC-expansion disorder, Friedreich ataxia (FRDA) (52) and in induced pluripotent stem cells derived from FRDA patients (53). However, MutSα protects against germ line expansion in the FRDA mouse (50) and against somatic expansion in a mouse model for the CTG/CAG-expansion disorder, myotonic dystrophy type 1 (54). Furthermore, it has no effect on germ line expansion in another model of the same disease (49) or in either germ line or somatic expansion in a mouse model of Huntington disease, which results from expansion of the same repeat (51). Why different mouse models differ with respect to the role played by MutSβ and MutSα remains to be seen.
Analysis of the distribution of repeat contractions in Msh3+/+ animals revealed the existence of two different contraction size classes, one involving the loss of just 1–2 repeats and a second class involving the loss of a much larger number of repeats (Fig. 3). As Msh3−/− animals have a distribution of contractions that is strongly shifted toward alleles that have lost fewer repeats, our data suggest that MutSβ is specifically involved in the generation of large contractions. The fact that MutSβ preferentially affects larger contractions but does not eliminate expansions altogether lends support to the idea that there are two different contraction mechanisms operating in the FXD mouse model, one that is MutSβ independent and one that is MutSβ dependent. It may be that the MutSβ-dependent contractions represent one outcome of the same process that gives rise to expansions. This would be of interest because it would indicate that while the mechanism responsible for expansion is strongly expansion biased, it can also generate contractions, albeit at lower frequency. This would have interesting implications for the underlying mechanism. The nature of the MutSβ-independent contraction pathway remains unclear.
To date there have been no other reports of MutSβ-dependent contractions in any other Repeat Expansion Disease mouse model, although the loss of MutSβ was shown to decrease contractions in repeat-containing minigene construct inserted into a human fibrosarcoma cell line (55). However, as contractions occur at such low frequencies in mice WT for MutSβ, it may be that larger studies in these models are needed to fully address this possibility. Whether MutSβ-dependent expansions and contractions arise via the same pathway in the FXD mouse model is not known. Our data showing that MutSβ binds well to both strands of the FX repeat suggest the possibility that both strands are potential substrates for MutSβ-dependent events (Fig. 4).
Our evidence that the kinetics of ATP hydrolysis is altered by loop-outs of either strand suggests that the conformation of MutSβ on binding to the loop-outs may differ from its conformation on binding to a canonical MMR substrate (Fig. 5). It has been suggested that the substrate-dependent regulation of the ATP hydrolytic cycle is critical for differentiating between different repair pathways (42–44). If this is in fact so, it may be that the altered kinetics of ATP hydrolysis relatively to a typical MMR substrate reflects the involvement of a different repair pathway in the generation of expansions and contractions. The effect of MutSβ on CCG-hairpin stability suggests that one way that MutSβ could act to promote both expansions and contractions is by increasing the longevity of the hairpins (Table 2), perhaps giving time for these structures to be channeled into an alternative pathway that generates expansions. We have previously shown that Polβ, a key polymerase in the base excision repair pathway, is also important for repeat expansion (56). However, while a hypomorphic POLΒ mutation decreases the expansion frequency, our data suggest that it does not decrease the frequency of large contractions. One possibility is that MutSβ acts upstream of Polβ to bind the substrate for instability, channeling some bound substrates into the Polβ pathway to generate expansions while diverting others into a different pathway that results in contractions. The nature of the MutSβ-independent contraction pathway is unknown.
Materials and Methods
Mouse maintenance
The generation of the FXD PM mice was described previously (2). The Msh3+/− mice were generated previously (57,58), and cryopreserved embryos were obtained from the NCI Mouse Repository (Frederick, MD). Live born pups were generated from these embryos by implantation into the oviduct of pseudopregnant recipients using standard procedures. Multiple breeding pairs were set up from Msh3 heterozygous parents to generate the appropriate genotypes. Msh3+/+ littermates from these crosses were used as controls. Mice were maintained in accordance with the guidelines of the NIDDK Animal Care and Use Committee and with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23, revised 1996).
Genotyping and analysis of repeat number
Genomic DNA from mouse tails was extracted using KAPA Mouse Genotyping Kit (KAPA Biosystems, Wilmington, MA). Genomic DNA from other tissues was extracted using a Maxwell®16 Mouse tail DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. Msh3 genotyping was carried out as described elsewhere (http://mouse.ncifcrf.gov/protocols.asp?ID=01XA3&p_allele=Msh3%3Ctm1Rak%3E&prot_no=1). The presence of the PM allele and its repeat number were determined using a fluorescent PCR assay and FraxM4 and FraxM5 primer pair as described previously (3). The somatic instability index (SII) was calculated from the GeneMapper profiles of DNA from different organs as previously described (3,35) and used to evaluate the extent of somatic expansion in adult mice. Fisher's exact test and the calculation of 95% confidence intervals were carried out using the GraphPad QuickCalcs website (http://www.graphpad.com/quickcalcs). Hartigans′ dip test was implemented using the dip.test command in the R diptest library.
Purification of human MutSβ
Human MutSβ was purified as described previously (45). Briefly, bacmids containing a region coding for His8-MBP-tagged human MSH2 and MSH3 were transfected into SF-9 cells in Grace's Insect Cell Medium using Cellfectin II (Life Technologies, Grand Island, NY) according to the manufacturer's instructions. MSH2 and MSH3 were then co-expressed by infecting Hi5 insect cells with both of the viruses. The cells were harvested at 48 h after infection, lysed in Buffer A [25 mM Hepes pH 8.0, 1 M NaCl, 30 mm imidazole, 10% (v/v) glycerol and 1 mm Tris (2-carboxyethyl)phosphine hydrochloride (TCEP) and MutSβ affinity purified using a Ni2+ affinity column (GE Healthcare, Piscataway, NJ)]. The proteins were eluted with a step gradient of 30% (v/v) Buffer B (Buffer A+300 mm imidazole). The (His)8-MBP tags were cleaved off using PreScission protease (GE Healthcare) and removed by passage over Amylose resin. The proteins were further purified using Heparin, Mono Q (GE Healthcare) and Superdex-200 columns. Proteins were stored in a buffer containing 25 mm HEPES pH 8.0, 100 mm KCl, 5 mm MgCl2, 0.1 mm EDTA, 5–30% (v/v) glycerol and 1 mm TCEP at −20°C. The purity of the preparation was checked by SDS–PAGE gel electrophoresis of the Mono Q and Superdex-200 fractions. No contaminating proteins were seen in the MutSβ preparation, and equivalent amounts of MSH2 and MSH3 are present (Supplementary Material, Fig. S1).
EMSA analysis of MutSβ binding to CGG, CCG, CTG and CAG-repeats
The oligonucleotides used for this study are listed in Table 1 and were annealed as described previously (41). The bottom strand oligonucleotide in each case (DuplexBS) was labeled at the 5′ end with biotin (Integrated DNA Technology, Coralville, IA). The binding reactions were carried out using the Gelshift™ chemiluminescent EMSA kit (Active Motif, Carlsbad, CA) according to the manufacturer's instructions. Briefly, purified human MutSβ (0–4 nm) was incubated with 2 fmoles of the duplexed oligonucleotides in a buffer containing 25 mm HEPES pH 8.0, 100 mm NaCl, 5 mm MgCl2, 0.1 mm EDTA, 4% (v/v) glycerol, 2 mm TCEP and 0.2 mg/ml BSA and incubated for 10 min at room temperature. The complexes were resolved on a 6% (w/v) DNA-Retardation gel (Life Technologies) in 0.5× TBE at 4°C. The DNA and the DNA–protein complexes were transferred to a nylon membrane, UV cross-linked using a Stratalinker (formerly Stratagene, now Agilent Technologies, Santa Clara, CA). The membrane was then blocked, incubated with Streptavidin-HRP conjugate and the DNA detected as per the kit protocol.
MutSβ ATPase assay
The MutSβ ATPase activity was assayed in 15 µl of a buffer containing 25 mm HEPES (pH 8.0), 100 mm KCl, 2 mm TCEP, 5% glycerol, 5 mm MgCl2, and 100 nm or 200 nm human MutSβ and 400 nm or 300 nm of the indicated oligonucleotide substrates. Kinetic data were obtained by varying the ATP concentration from 10 to 80 μM containing 50 fmol of α-32P-ATP (PerkinElmer, Waltham, MA) in each reaction. The reaction was incubated at 37°C for 15–30 min and terminated by addition of an equal volume of 50 mm EDTA. One microliter of each reaction mix was spotted on a PEI-cellulose TLC plate (Grace Davison Discovery Sciences, Albany, OR). Labeled ATP and ADP were separated by developing the TLC plate in 0.75 M KH2PO4 and visualized by storage phosphor autoradiography in a Typhoon TRIO Imager (GE Healthcare). ImageQuant TL software (GE Healthcare) was used to quantify the products, and GraphPad Prism (GraphPad Software, La Jolla, CA) was used to plot the graphs and calculate Km and kcat values.
Thermal analysis of the CCG-repeats in the presence of MutSβ
Fluorescently labeled (CCG)10 oligonucleotide was resuspended in a buffer containing 25 mm HEPES at pH 7.3, 25 mm KCl, 1 mm MgCl2, 0.1 mm EDTA, 4% (v/v) glycerol, 2 mm TCEP and 0.2 mg/ml BSA. The oligonucleotide (90 nm) was heated to 95°C for 5 min and then allowed to cool to room temperature to permit hairpin formation. Human MutSβ or BSA was added to a concentration of 360 nm as indicated. Thermal denaturation was monitored by measuring the change in ROX™ fluorescence at 608 nm in a StepOnePlus™ real-time PCR machine (Applied Biosystems, Carlsbad, CA) with a heating rate of 0.6°C/min. The data reported represent the average of three replicates. The thermodynamic parameters were derived from the melting curve using a two-state model (closed and open states).
Supplementary Material
Funding
The work described in this manuscript was funded by a grant from the Intramural Program of the NIDDK to K.U. (DK057808-07).
Supplementary Material
Acknowledgements
The Usdin lab would like to acknowledge all the hard work by the people that take care of the mice used in this study. Without their help, this work would not have been possible. We would also like to thank Dr Bruce Hayward for his computational assistance.
Conflict of Interest statement. None declared.
References
- 1.Loesch D., Hagerman R. (2012) Unstable mutations in the FMR1 gene and the phenotypes. Adv. Exp. Med. Biol., 769, 78–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Entezam A., Biacsi R., Orrison B., Saha T., Hoffman G.E., Grabczyk E., Nussbaum R.L., Usdin K. (2007) Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene, 395, 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lokanga R.A., Entezam A., Kumari D., Yudkin D., Qin M., Smith C.B., Usdin K. (2013) Somatic expansion in mouse and human carriers of fragile X premutation alleles. Hum. Mutat., 34, 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X.N., Usdin K. (2014) Gender and cell-type-specific effects of the transcription-coupled repair protein, ERCC6/CSB, on repeat expansion in a mouse model of the fragile X-related disorders. Hum. Mutat., 35, 341–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petek E., Kroisel P.M., Schuster M., Zierler H., Wagner K. (1999) Mosaicism in a fragile X male including a de novo deletion in the FMR1 gene. Am. J. Med. Genet., 84, 229–232. [PubMed] [Google Scholar]
- 6.Schmucker B., Seidel J. (1999) Mosaicism for a full mutation and a normal size allele in two fragile X males. Am. J. Med. Genet., 84, 221–225. [PubMed] [Google Scholar]
- 7.Todorov T., Todorova A., Kirov A., Dimitrov B., Carvalho R., Nygren A.O., Boneva I., Mitev V. (2009) Fragile X mosaic male full mutation/normal allele detected by PCR/MS-MLPA. BMJ Case Rep., 2009, doi:10.1136/bcr.06.2008.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia Arocena D., de Diego Y., Oostra B.A., Willemsen R., Mirta Rodriguez M. (2000) A fragile X case with an amplification/deletion mosaic pattern. Hum. Genet., 106, 366–369. [DOI] [PubMed] [Google Scholar]
- 9.de Graaff E., de Vries B.B., Willemsen R., van Hemel J.O., Mohkamsing S., Oostra B.A., van den Ouweland A.M. (1996) The fragile X phenotype in a mosaic male with a deletion showing expression of the FMR1 protein in 28% of the cells. Am. J. Med. Genet., 64, 302–308. [DOI] [PubMed] [Google Scholar]
- 10.Fan H., Booker J.K., McCandless S.E., Shashi V., Fleming A., Farber R.A. (2005) Mosaicism for an FMR1 gene deletion in a fragile X female. Am. J. Med. Genet. A, 136, 214–217. [DOI] [PubMed] [Google Scholar]
- 11.Tabolacci E., Pomponi M.G., Pietrobono R., Chiurazzi P., Neri G. (2008) A unique case of reversion to normal size of a maternal premutation FMR1 allele in a normal boy. Eur. J. Hum. Genet., 16, 209–214. [DOI] [PubMed] [Google Scholar]
- 12.Gasteiger M., Grasbon-Frodl E., Neitzel B., Kooy F., Holinski-Feder E. (2003) FMR1 gene deletion/reversion: a pitfall of fragile X carrier testing. Genet Test, 7, 303–308. [DOI] [PubMed] [Google Scholar]
- 13.Govaerts L.C., Smit A.E., Saris J.J., VanderWerf F., Willemsen R., Bakker C.E., De Zeeuw C.I., Oostra B.A. (2007) Exceptional good cognitive and phenotypic profile in a male carrying a mosaic mutation in the FMR1 gene. Clin. Genet., 72, 138–144. [DOI] [PubMed] [Google Scholar]
- 14.Grasso M., Faravelli F., Lo Nigro C., Chiurazzi P., Sperandeo M.P., Argusti A., Pomponi M.G., Lecora M., Sebastio G.F., Perroni L. et al. (1999) Mosaicism for the full mutation and a microdeletion involving the CGG repeat and flanking sequences in the FMR1 gene in eight fragile X patients. Am. J. Med. Genet., 85, 311–316. [DOI] [PubMed] [Google Scholar]
- 15.Mila M., Castellvi-Bel S., Sanchez A., Lazaro C., Villa M., Estivill X. (1996) Mosaicism for the fragile X syndrome full mutation and deletions within the CGG repeat of the FMR1 gene. J. Med. Genet., 33, 338–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Usdin K., Woodford K.J. (1995) CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucl. Acids Res., 23, 4202–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu A., Dill J., Mitas M. (1995) The purine-rich trinucleotide repeat sequences d(CAG)15 and d(GAC)15 form hairpins. Nucl. Acids Res., 23, 4055–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitas M., Yu A., Dill J., Haworth I.S. (1995) The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn. Ganti base pairs. Biochemistry, 34, 12803–12811. [DOI] [PubMed] [Google Scholar]
- 19.Mitas M., Yu A., Dill J., Kamp T.J., Chambers E.J., Haworth I.S. (1995) Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucl. Acids Res., 23, 1050–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu A., Barron M.D., Romero R.M., Christy M., Gold B., Dai J., Gray D.M., Haworth I.S., Mitas M. (1997) At physiological pH, d(CCG)15 forms a hairpin containing protonated cytosines and a distorted helix. Biochemistry, 36, 3687–3699. [DOI] [PubMed] [Google Scholar]
- 21.Gacy A.M., Goellner G., Juranic N., Macura S., Mcmurray C.T. (1995) Trinucleotide repeats that expand in human-disease form hairpin structures in-vitro. Cell, 81, 533–540. [DOI] [PubMed] [Google Scholar]
- 22.Gacy A.M., Goellner G.M., Spiro C., Chen X., Gupta G., Bradbury E.M., Dyer R.B., Mikesell M.J., Yao J.Z., Johnson A.J. et al. (1998) GAA instability in Friedreich's Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell, 1, 583–593. [DOI] [PubMed] [Google Scholar]
- 23.Fry M., Loeb L.A. (1994) The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA, 91, 4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadel Y., Weisman-Shomer P., Fry M. (1995) The fragile X syndrome single strand d(CGG)n nucleotide repeats readily fold back to form unimolecular hairpin structures. J. Biol. Chem., 270, 28970–28977. [DOI] [PubMed] [Google Scholar]
- 25.Saha T., Usdin K. (2001) Tetraplex formation by the progressive myoclonus epilepsy type-1 repeat: implications for instability in the repeat expansion diseases. FEBS Lett., 491, 184–187. [DOI] [PubMed] [Google Scholar]
- 26.Sket P., Pohleven J., Kovanda A., Stalekar M., Zupunski V., Zalar M., Plavec J., Rogelj B. (2015) Characterization of DNA G-quadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging, 36, 1091–1096. [DOI] [PubMed] [Google Scholar]
- 27.Latham G.J., Coppinger J., Hadd A.G., Nolin S.L. (2014) The role of AGG interruptions in fragile X repeat expansions: a twenty-year perspective. Front Genet., 5, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yrigollen C.M., Martorell L., Durbin-Johnson B., Naudo M., Genoves J., Murgia A., Polli R., Zhou L., Barbouth D., Rupchock A. et al. (2014) AGG interruptions and maternal age affect FMR1 CGG repeat allele stability during transmission. J. Neurodev. Disord., 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolin S.L., Glicksman A., Ersalesi N., Dobkin C., Brown W.T., Cao R., Blatt E., Sah S., Latham G.J., Hadd A.G. (2015) Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet. Med., 17, 358–364. [DOI] [PubMed] [Google Scholar]
- 30.Mirkin S.M. (2007) Expandable DNA repeats and human disease. Nature, 447, 932–940. [DOI] [PubMed] [Google Scholar]
- 31.Lokanga R.A., Zhao X.N., Entezam A., Usdin K. (2014) X inactivation plays a major role in the gender bias in somatic expansion in a mouse model of the fragile X-related disorders: implications for the mechanism of repeat expansion. Hum. Mol. Genet., 23, 4985–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grasso M., Boon E.M., Filipovic-Sadic S., van Bunderen P.A., Gennaro E., Cao R., Latham G.J., Hadd A.G., Coviello D.A. (2014) A novel methylation PCR that offers standardized determination of FMR1 methylation and CGG repeat length without southern blot analysis. J. Mol. Diagn., 16, 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lokanga R.A., Zhao X.N., Usdin K. (2014) The mismatch repair protein MSH2 is rate limiting for repeat expansion in a fragile X premutation mouse model. Hum. Mutat., 35, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kovtun I.V., McMurray C.T. (2008) Features of trinucleotide repeat instability in vivo. Cell Res., 18, 198–213. [DOI] [PubMed] [Google Scholar]
- 35.Lee J.M., Zhang J., Su A.I., Walker J.R., Wiltshire T., Kang K., Dragileva E., Gillis T., Lopez E.T., Boily M.J. et al. (2010) A novel approach to investigate tissue-specific trinucleotide repeat instability. BMC Syst. Biol., 4, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X.N., Usdin K. (2015) The transcription-coupled repair protein ERCC6/CSB also protects against repeat expansion in a mouse model of the fragile X premutation. Hum. Mutat., 36, 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller M.J., Yuan B.Z. (1997) Semiautomated resolution of overlapping stutter patterns in genomic microsatellite analysis. Anal. Biochem., 251, 50–56. [DOI] [PubMed] [Google Scholar]
- 38.Walsh P.S., Fildes N.J., Reynolds R. (1996) Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucl. Acids Res., 24, 2807–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinde D., Lai Y., Sun F., Arnheim N. (2003) Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucl. Acids Res., 31, 974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartigan J.A., Hartigan P.M. (1985) The Dip Test of Unimodality. Ann. Stat., 13, 70–84. [Google Scholar]
- 41.Owen B.A., Yang Z., Lai M., Gajec M., Badger J.D. II, Hayes J.J., Edelmann W., Kucherlapati R., Wilson T.M., McMurray C.T. (2005) (CAG)(n)-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol., 12, 663–670. [DOI] [PubMed] [Google Scholar]
- 42.Kumar C., Eichmiller R., Wang B., Williams G.M., Bianco P.R., Surtees J.A. (2014) ATP binding and hydrolysis by Saccharomyces cerevisiae Msh2-Msh3 are differentially modulated by mismatch and double-strand break repair DNA substrates. DNA Repair, 18, 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang W.H., Coats J.E., Majka J., Hura G.L., Lin Y., Rasnik I., McMurray C.T. (2011) Conformational trapping of mismatch recognition complex MSH2/MSH3 on repair-resistant DNA loops. Proc. Natl Acad. Sci. USA, 108, E837–E844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owen B.A., W H.L., McMurray C.T. (2009) The nucleotide binding dynamics of human MSH2-MSH3 are lesion dependent. Nat. Struct. Mol. Biol., 16, 550–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S., Gellert M., Yang W. (2012) Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops. Nat. Struct. Mol. Biol., 19, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usdin K. (1998) NGG-triplet repeats form similar intrastrand structures: implications for the triplet expansion diseases. Nucl. Acids Res., 26, 4078–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amrane S., Mergny J.L. (2006) Length and pH-dependent energetics of (CCG)n and (CGG)n trinucleotide repeats. Biochimie, 88, 1125–1134. [DOI] [PubMed] [Google Scholar]
- 48.Kovtun I.V., Goellner G., McMurray C.T. (2001) Structural features of trinucleotide repeats associated with DNA expansion. Biochem. Cell Biol., 79, 325–336. [PubMed] [Google Scholar]
- 49.Foiry L., Dong L., Savouret C., Hubert L., te Riele H., Junien C., Gourdon G. (2006) Msh3 is a limiting factor in the formation of intergenerational CTG expansions in DM1 transgenic mice. Hum. Genet., 119, 520–526. [DOI] [PubMed] [Google Scholar]
- 50.Ezzatizadeh V., Pinto R.M., Sandi C., Sandi M., Al-Mahdawi S., Te Riele H., Pook M.A. (2012) The mismatch repair system protects against intergenerational GAA repeat instability in a Friedreich ataxia mouse model. Neurobiol. Dis., 46, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dragileva E., Hendricks A., Teed A., Gillis T., Lopez E.T., Friedberg E.C., Kucherlapati R., Edelmann W., Lunetta K.L., MacDonald M.E. et al. (2009) Intergenerational and striatal CAG repeat instability in Huntington's disease knock-in mice involve different DNA repair genes. Neurobiol. Dis., 33, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bourn R.L., De Biase I., Pinto R.M., Sandi C., Al-Mahdawi S., Pook M.A., Bidichandani S.I. (2012) Pms2 suppresses large expansions of the (GAA.TTC)n sequence in neuronal tissues. PLoS One, 7, e47085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du J., Campau E., Soragni E., Ku S., Puckett J.W., Dervan P.B., Gottesfeld J.M. (2012) Role of mismatch repair enzymes in GAA.TTC triplet-repeat expansion in Friedreich ataxia induced pluripotent stem cells. J. Biol. Chem., 287, 29861–29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Broek W.J., Nelen M.R., Wansink D.G., Coerwinkel M.M., te Riele H., Groenen P.J., Wieringa B. (2002) Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet., 11, 191–198. [DOI] [PubMed] [Google Scholar]
- 55.Lin Y., Wilson J.H. (2009) Diverse effects of individual mismatch repair components on transcription-induced CAG repeat instability in human cells. DNA Repair, 8, 878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lokanga R.A., Senejani A.G., Sweasy J.B., Usdin K. (2015) Heterozygosity for a hypomorphic polbeta mutation reduces the expansion frequency in a mouse model of the fragile x-related disorders. PLoS Genet, 11, e1005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edelmann W., Yang K., Umar A., Heyer J., Lau K., Fan K., Liedtke W., Cohen P.E., Kane M.F., Lipford J.R. et al. (1997) Mutation in the mismatch repair gene Msh6 causes cancer susceptibility. Cell, 91, 467–477. [DOI] [PubMed] [Google Scholar]
- 58.Edelmann W., Umar A., Yang K., Heyer J., Kucherlapati M., Lia M., Kneitz B., Avdievich E., Fan K., Wong E. et al. (2000) The DNA mismatch repair genes Msh3 and Msh6 cooperate in intestinal tumor suppression. Cancer Res., 60, 803–807. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.