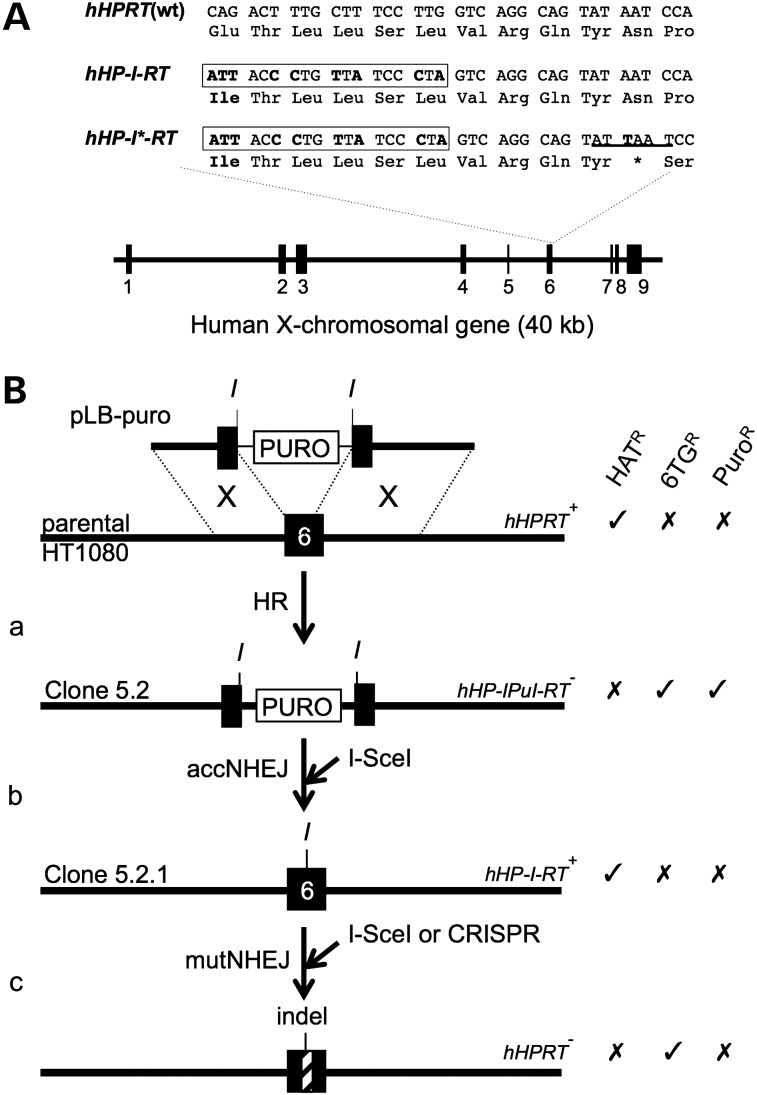
Figure 3.
Generating and using I-SceI-sensitive hHPRT alleles. (A) Structure of parental and I-SceI-sensitive hHPRT alleles. Numbered black boxes represent exons. Partial nucleotide and amino-acid sequences for exon 6 are shown for the WT allele and for alleles modified to carry an I-SceI site (boxed), with (hHP-I*-RT) or without (hHP-I-RT) an adjacent stop codon. Altered residues are shown in bold. An AseI site in hHP-I*-RT is underlined. (B) Strategy for generating I-SceI-sensitive alleles and using them for accNHEJ and mutNHEJ assays. (a) Disruption of exon 6 by GT with pLB-puro. (b) Basis of accNHEJ assay. An I-SceI-induced DSB repaired by accNHEJ generates a functional I-SceI-sensitive allele. The frequency of resulting HATR colonies measures accNHEJ. (c) Basis of mutNHEJ assay. An I-SceI-induced DSB is repaired by mutNHEJ to generate an indel (hatched box). The frequency of resulting 6TGR colonies measures mutNHEJ. Exon 6 (black box) and adjacent intronic DNA of the chromosomal HPRT locus (long lines) are shown. Targeting construct pLB-puro is shown with its puromycin-resistance cassette (PURO, white box) and its 2.7 and 3.1 kb arms aligned with homologous chromosomal regions (dotted lines) to allow HR (X). Sites for I-SceI (I) are shown. Alleles are labelled with their names (right) and the names of representative host cell lines (left). Resistance (✓) or sensitivity (✗) to key selective agents is indicated.