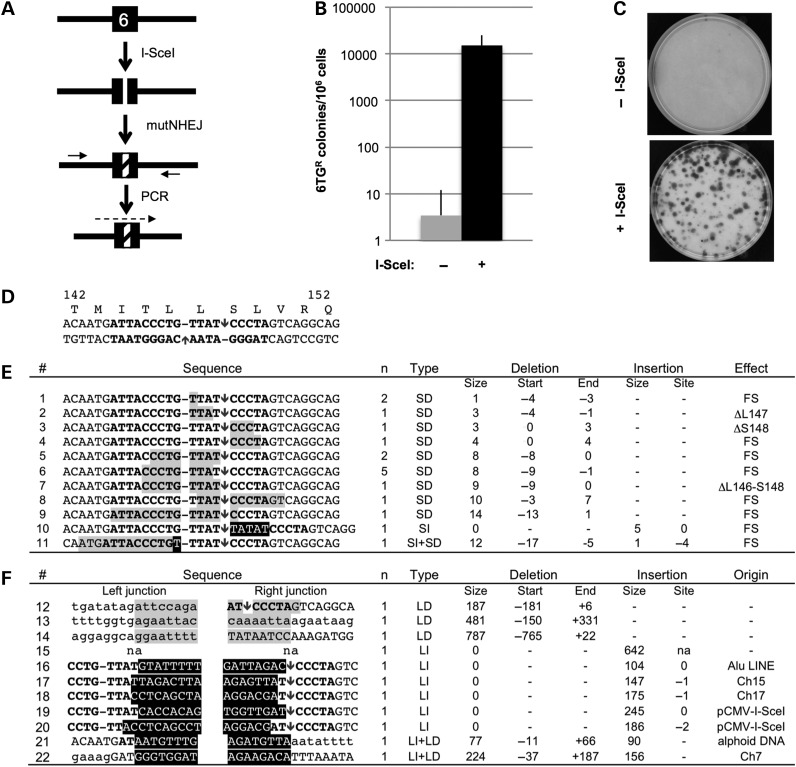
Figure 4.
Assays for I-SceI-induced mutNHEJ and DNA sequence analyses of resulting indels in HPRT exon 6. (A) HPRT exon 6 in clone 5.2.1 is cleaved by I-SceI and repaired by mutNHEJ to generate 6TGR clones carrying exon 6 indels (hatched); these are amplified with flanking primers (short arrows) and sequenced (dashed arrow). (B) Frequency of 6TGR clones generated from clone 5.2.1 lipofected with I-SceI expression plasmid or a vector control (mean and SD for six experiments are shown). Transfection was by lipofectamine 2000, but similar results were obtained with Fugene. (C) Example of 6TGR colonies forming in the experiment in B. Petri dishes (10 cm diameter) were seeded with 105 cells before selecting in 6TG. (D) Part of the exon 6 DNA sequence in clone 5.2.1 is shown aligned with encoded amino acids 142–152. The recognition site for I-SceI is shown (bold) with positions of staggered nicks it makes (arrows) and un-cleaved phosphodiester bonds opposite each nick (hyphens). (E) Small indels. Coding strand DNA sequences of 11 indels identified in seventeen 6TGR clones are shown with deleted and inserted residues highlighted in grey and black, respectively. The number of clones for each indel is shown (n). Positions of indels are summarized with residues numbered relative to the coding strand nick. Micro-homologies likely to have mediated deletion formation are underlined. Indel types: small and large deletions (SD, LD), or insertions (SI, LI). The predicted effects on protein coding are categorized as frame-shifts (FS) or in-frame deletions (Δ). (F) Large indels. Sequences of 11 junction regions are shown with DNA represented as in (B). Except for insertions, upper and lower case letters represent exon 6 and intronic residues, respectively. Type and position numbers of indels are summarized as in (B). The origin of the inserted DNA is indicated. na, no sequence available.