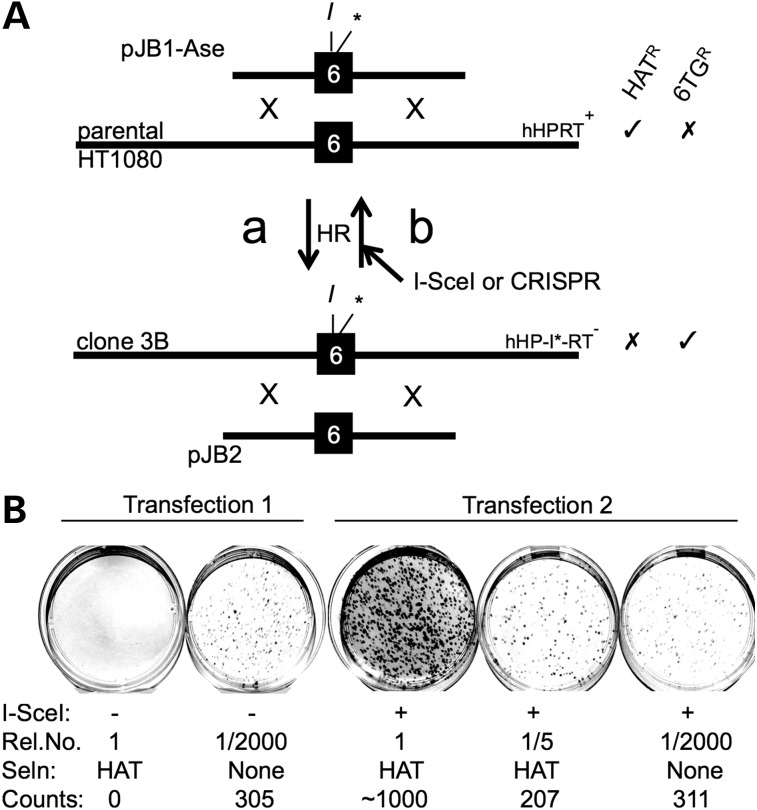
Figure 5.
Generating and using I-SceI-sensitive hHPRT+ alleles for gc/HR assays. (A) DNA is represented as in Figure 3. (a) HR between a wt HPRT allele and the targeting construct pJB1-AseI was used to introduce an I-SceI site and adjacent stop codon (*) into exon 6. The resulting cells (e.g. clone 3B) were selected in 6TG. (b) Basis of assays for gene correction by HR (gc/HR). Clone 3B cells are transfected with a repair template (pJB2) and nuclease expression plasmid(s). HR with transfected pJB2 regenerates a functional exon 6 and the frequency of resulting HATR colonies measures gc/HR. (B) Example of one experiment to estimate gc/HR frequencies. Clone 3B cells were co-transfected with pJB2 and either vector DNA (Transfection 1) or I-SceI expression plasmid (Transfection 2), and the indicated relative numbers (Rel. No.) of cells were placed in petri dishes to select in HAT for gene correction events or to determine plating efficiencies (no selection). Colonies appearing after ∼10 days were counted and used to estimate gc/HR frequencies of <0.00016% (Transfection 1) and 0.17% (Transfection 2).