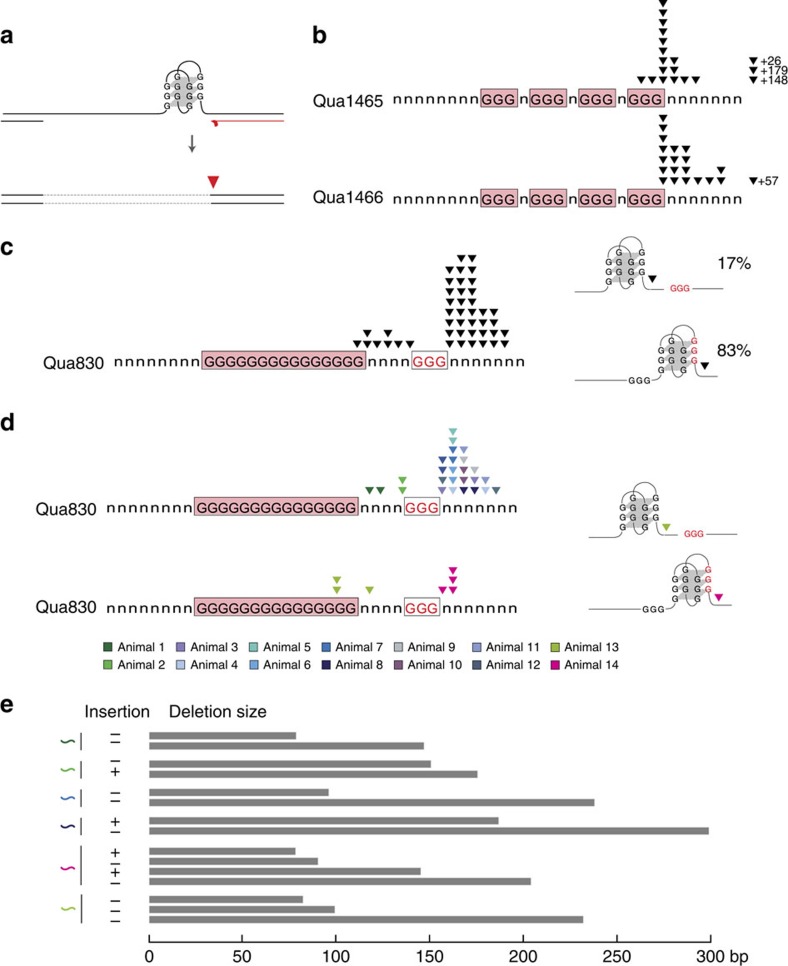
Figure 3. Signatures of inheritable G-quadruplex configurations.
(a) Current model clarifying the correlation between the genomic position of a G4 motif and the 3′-junction of the cognate G-quadruplex-induced deletion: 3′-deletion junctions (red triangle) are defined by the halted progression of nascent strand synthesis at a G-quadruplex fold (red line). (b) Spectra of 3′-deletion junctions at indicated G4 loci. Each black triangle represents a 3′-deletion junction independently derived from single animals. (c) Bimodal spectrum of 3′-deletion junctions at Qua830. Black triangles represent 3′-deletion junctions independently isolated from individual animals. Illustrations on the right portray potential G-quadruplex configurations at Qua830 and the relative frequency of deletions, which, based on the position of their 3′-junctions, are attributed to that particular configuration. (d) Results of 3′-deletion junction analysis at Qua830 in single dog-1-deficient animals. The 3′-deletion junctions identified in 1% lysate fractions of the same individual are colour coded as indicated. Illustrations on the right portray G-quadruplex configurations at Qua830 and the position of recurrent 3′-deletion junctions identified in animal 13 and 14. (e) Graphical illustration of the G4 deletions described and depicted in d. The presence or absence of templated insertions is indicated (+/−, respectively).