Fig. 5.
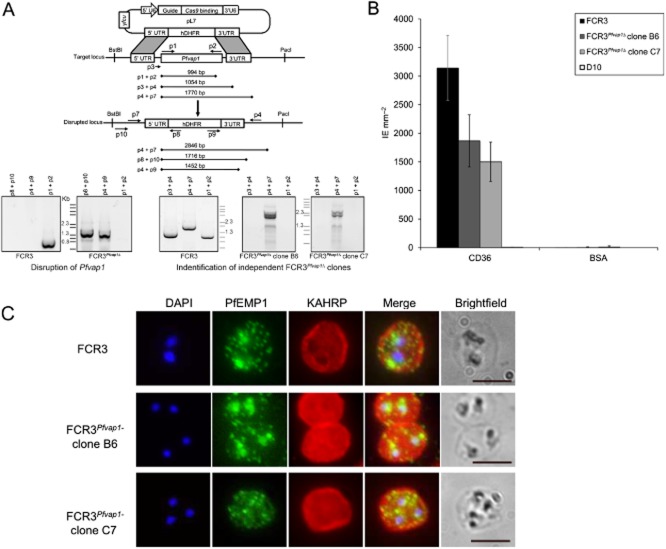
FCR3Pfvap1Δ knockout parasites show a reduction in cytoadhesion.A. Diagram depicting the cas9/CRISPR strategy (Ghorbal et al., 2014) used for disruption of Pfvap1 in FCR3 parasites. Only the pL7 plasmid carrying the Pfvap1-specific guide RNA and homology boxes is shown. The position of primers is shown (arrows) and each primer (p) is numbered. The expected amplicon sizes for PCR verification are shown. Gene replacement with hDHFR was verified by PCR (left). In FCR3Pfvap1Δ parasites, part of the 5′UTR sequence is lost (p3), the absence of this sequence used to select two KO clones (right).B. Binding of FCR3Pfvap1Δ knockout clones panned on CD36. A 40% reduction in binding was observed in FCR3Pfvap1Δ clone B6 (dark grey). Binding to CD36 was reduced by 50% in the C7 clone (light grey). Mean binding of IE mm−2 to purified CD36 and BSA is shown. Error bars = o standard deviation, n = 3 replicates.C. FCR3Pfvap1Δ parasites export PfEMP1 and KAHRP in a manner comparable with wild-type FCR3 parasites, suggesting that there is no impairment or retention of these proteins in mutant parasites. Scale bar = 5 μm.