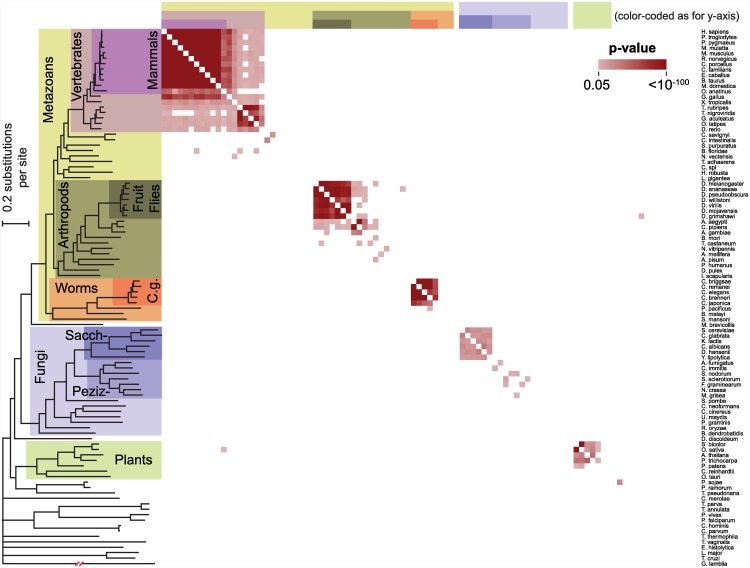
Fig 2. Conservation of Puf3 RNA targets across eukaryotes.
A heatmap displaying whether the Puf3 binding sequence UGUA[ACU]AUA is shared by orthologs in a pair of species. A significant p-value is displayed as a colored square and was determined by the hypergeometric test. Color is used only if the Puf3 motif ranks in the top 1% relative to all motif permutations (Materials and Methods). (The heatmap is not fully symmetrical since the orthology assignments are not necessarily reciprocal.) The phylogeny shown was inferred using a maximum-likelihood approach from a concatenated alignment of 53 protein sequences (Materials and Methods). "Sacch-" refers to Saccharomycotina fungi, "Peziz-" to Pezizomycotina fungi, and "C.g." to the Caenorhabditis genus of worms. The break noted in red corresponds to 0.5 substitutions per site in the branch to Giardia lamblia. p-Values and ranks of the Puf3 motif against its permutations can be found in S9 Dataset.