Abstract
The neural crest is an evolutionary novelty that fostered the emergence of vertebrate anatomical innovations such as the cranium and jaws1. During embryonic development, multipotent neural crest cells are specified at the lateral borders of the neural plate before delaminating, migrating, and differentiating into various cell types. In invertebrate chordates (cephalochordates and tunicates), neural plate border cells express conserved factors such as Msx, Snail, and Pax3/7 and generate melanin-containing pigment cells2-4, a derivative of the neural crest in vertebrates. However, invertebrate neural plate border cells have not been shown to generate homologues of other neural crest derivatives. Thus, proposed models of neural crest evolution postulate vertebrate-specific elaborations on an ancestral neural plate border program, through acquisition of migratory capabilities and the potential to generate multiple cell types5-7. Here we show that a particular neuronal cell type in the tadpole larva of the tunicate Ciona intestinalis, the bipolar tail neuron, shares a set of features with neural crest-derived spinal ganglia neurons in vertebrates. Bipolar tail neuron precursors derive from caudal neural plate border cells, delaminate, and migrate along the paraxial mesoderm on either side of the neural tube, eventually differentiating into afferent neurons that form synaptic contacts with both epidermal sensory cells and motor neurons. We propose that the neural plate borders of the chordate ancestor already produced migratory peripheral neurons and pigment cells, and that the neural crest evolved through the acquisition of a multipotent progenitor regulatory state upstream of multiple, pre-existing neural plate border cell differentiation programs.
Progenitor cells that fulfill all the criteria defining the neural crest have not been observed outside vertebrates. These criteria include an embryonic origin at the lateral borders of the neural plate, epithelium-to-mesenchyme transition (EMT), migratory behavior, and the potential to differentiate into diverse cell types such as neurons, bone, cartilage, and pigment cells.
In cephalochordates (amphioxus) and the tunicates Halocynthia and Ciona, a subset of neural plate border cells deploy a conserved melanocyte-specific gene network but do not migrate away from the neural tube2-4. Instead, they contribute locally to pigmented photoreceptor organs. In Ciona, the pigment cell precursors undergo an epithelial-to-mesenchymal transition (EMT) and remain inside the neural tube lumen, but can be induced to exit the neural tube through targeted mis-expression of the mesenchyme-specific transcription factor Twist-related4. Migratory pigment cell precursors have also been reported in larvae of the tunicate Ecteinascidia turbinata8.
In contrast, invertebrate homologues of neural crest-derived neurons have so far proved elusive. In tunicates, various neurons arise from the neural plate borders, but homology to vertebrate neural crest-derived neurons has been denied because they do not migrate, remaining instead in the dorsal neural tube or in the epidermis9,10. Migratory sensory neurons have been described in cephalochordate embryos, but these arise from ventral epidermis, not the neural plate borders, and reinsert into the epidermis after migrating11.
The recently identified bipolar tail neurons (BTNs)12 of Ciona larvae form axon fascicles that extend along the length of the tail on either side of the neural tube (Fig. 1a). These neurons express the proneural basic helix-loop-helix transcription factor Neurogenin (Neurog, Fig. 1b) and the LIM-homeodomain factor Islet (Fig 1a). Vertebrate Neurog and Islet orthologues are involved in specifying various neuronal subtypes including neural crest-derived dorsal root ganglia neurons (DRGNs), which also have a bipolar or pseudounipolar morphology and transmit peripheral mechanosensory inputs to the central nervous system13. Ciona BTNs also express Asic, the orthologue of Acid-sensing ion channels (ASICs) 14 that modulate touch sensitivity in vertebrate DRGNs. These parallels prompted us to investigate the embryological origins of the BTNs.
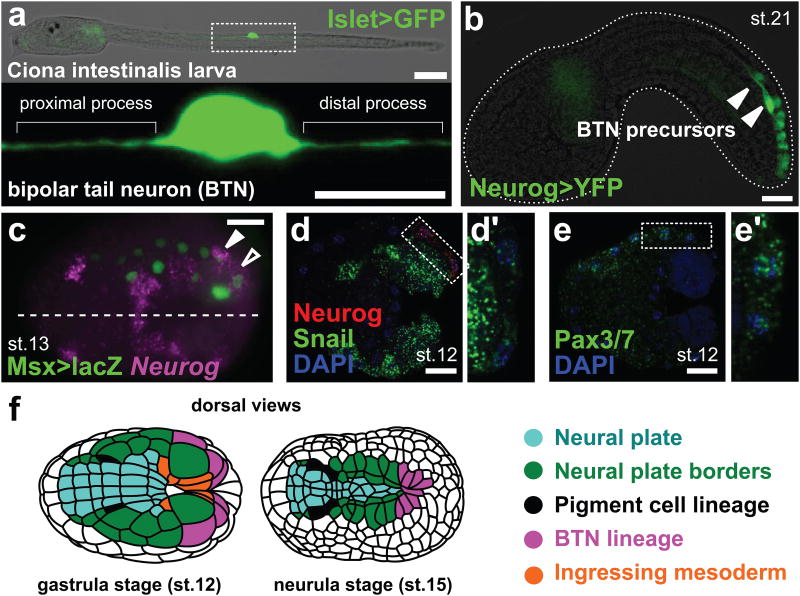
Figure 1. Bipolar tail neurons come from the borders of the neural plate.
a, Larva with a bipolar tail neuron (BTN) labeled by Islet BTN>unc-76∷eGFP (green). Lower panel: enlarged view of BTN above. Scale bars 75 μm (upper), 25 μm (lower). b, Migrating BTN precursors (arrowheads) labeled by Neurog b-line>unc-76∷Venus (green). Scale bar, 25 μm. c, In situ hybridization (ISH) for Neurog (magenta) in embryo electroporated with Msx>nls∷lacZ plasmid (immunolabeling of β-galactosidase in green). White arrowhead: Msx+/Neurog+ BTN progenitor. Black arrowhead: transient Neurog expression in BTN progenitor's sister cell (epidermal progenitor). Dashed line: midline. Scale bar, 25 μm. d, ISH for Neurog (red) and Snail (green). Scale bar, 25 μm. Box enlarged in d′ showing low levels of Snail expression in BTN progenitor. e, Pax3/7 ISH (green). Scale bar, 25 μm. Box enlarged in e′ showing Pax3/7 expression in BTN progenitor. f, Adapted illustration17 of embryos showing position of pigment cell and BTN progenitors (and their descendants) in the neural plate borders. Lateral views in a,b, dorsal views in c-f. Anterior to the left throughout. st. = stage.
We detected the earliest expression of Neurog at neurulation, in the caudal-most neural/epidermal boundary cells, which express the conserved neural plate border specification genes Msx15, Pax3/73, and Snail16 (Fig. 1c-f, Extended Data Fig. 1). During neurulation, these cells drive neural tube closure and their progeny eventually form the neural tube roof plate and dorsal epidermis midline17,18 (Fig. 1b, Extended Data Fig. 2). BTN progenitors are thus born from the caudal extensions of the lateral borders of the neural plate (Fig. 1f).
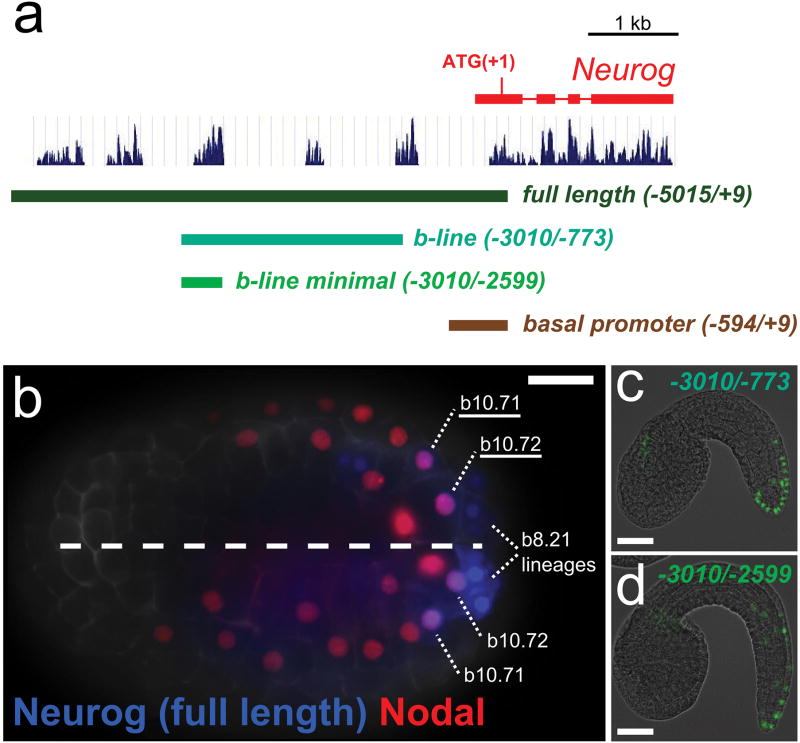
We isolated a Neurog cis-regulatory element that drives reporter gene expression in this caudal neural plate border region (Extended Data Fig. 3). Using this reporter, we determined that Neurog expression is progressively restricted and maintained in only two cells on each side of the bilaterally symmetric embryo, born during neural tube closure (Extended Data Fig. 2,4). We have named these the anterior (aBTN) and posterior (pBTN) BTN precursors. Shortly after completion of neural tube closure, BTN precursors delaminate and migrate anteriorly along the paraxial mesoderm on either side of the neural tube19 (Fig. 2a-f, Supplementary Video 1-3). This is evocative of vertebrate DRGN progenitors, which migrate through paraxial mesoderm situated lateral to the neural tube.
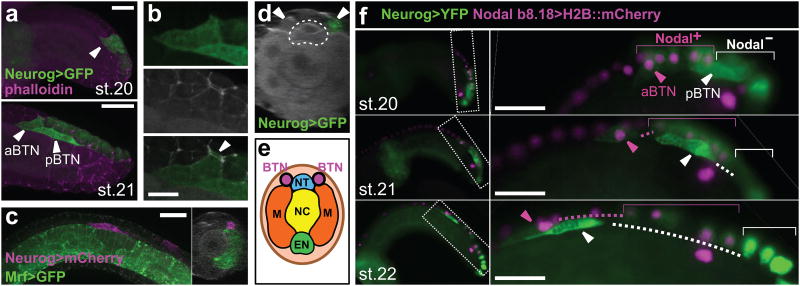
Figure 2. Bipolar tail neuron precursors delaminate and migrate.
a, Embryos electroporated with Neurog b-line>unc-76∷eGFP. Upper: Bipolar tail neuron (BTN) precursor (arrowhead) extending a lamellipodium. Lower: BTN precursors (anterior, aBTN and posterior, pBTN) delaminating. Scale bar, 25 μm. b, Enlarged view of aBTN in lower panel of a. Upper: UNC-76∷eGFP. Middle: Phalloidin. Lower: merged, arrowhead: part of aBTN still in the epithelium. Scale bar, 10 μm. c, Embryo with paraxial mesoderm labeled by Mrf>unc-76∷eGFP (green), BTN labeled by Neurog b-line>unc-76∷mCherry (magenta) and phalloidin counterstain. Scale bar 25 μm. Right: Cross sectioned 3D image of same embryo. Only the right side of the embryo was transfected. d, 3D slice of embryo showing BTNs (arrowheads) outside neural tube (dotted outline). Only the right side of the embryo was transfected. e, Diagram of d, showing BTNs relative to other tail tissues: neural tube (NT), notochord (NC), myoblasts (M), and endoderm (EN). f, Time series of different embryos co-electroporated with Neurog b-line>unc-76∷VenusYFP (green) and Nodal b8.18>H2B∷mCherry (magenta). Right panels are enlarged views from left. Dashed lines indicate displacement from clonally-related epidermal cells (indicated by color-coded brackets). Scale bars, 25 μm.
Double-labelling with a Nodal reporter revealed that BTNs arise from two adjacent but clonally distinct cell lineages (Fig. 2g and Extended Data Fig. 2). The pBTN arises from the tail tip (b8.21 lineage)10 and migrates to meet the b8.18-derived aBTN as it delaminates (Fig. 2a,f). Together, they continue their migration as a chain of two cells.
Neurog expression distinguishes the BTNs from the caudal epidermal sensory neurons (CESNs), which remain at the dorsal midline and are specified instead by an Atonal homolog (Atoh)-dependent regulatory program10,20. We found that the onset of Neurog expression requires MAPK/ERK signaling (Fig. 3a,b). However, later inhibition of MAPK/ERK resulted in the upregulation of Neurog in non-neural cells of the lineage, converting these into supernumerary BTNs (Fig. 3c-e and Extended Data Fig. 4). In contrast, perturbing Delta/Notch signaling did not alter BTN specification or differentiation (Extended Data Fig. 5). Overexpression of Neurog also induced ectopic migratory Asic+ BTN precursors (Fig. 3f,g), while BTNs were abolished through expression of a dominant repressor form of Neurog (Neurog∷WRPW, Fig. 3h). In all cases, induced supernumerary BTN precursors migrated as an expanded chain of cells (Fig. 3e,g). These data indicate that sustained Neurog expression in caudal neural plate border cells is controlled by MAPK/ERK signaling and is necessary and sufficient for BTN specification, migration, and differentiation.
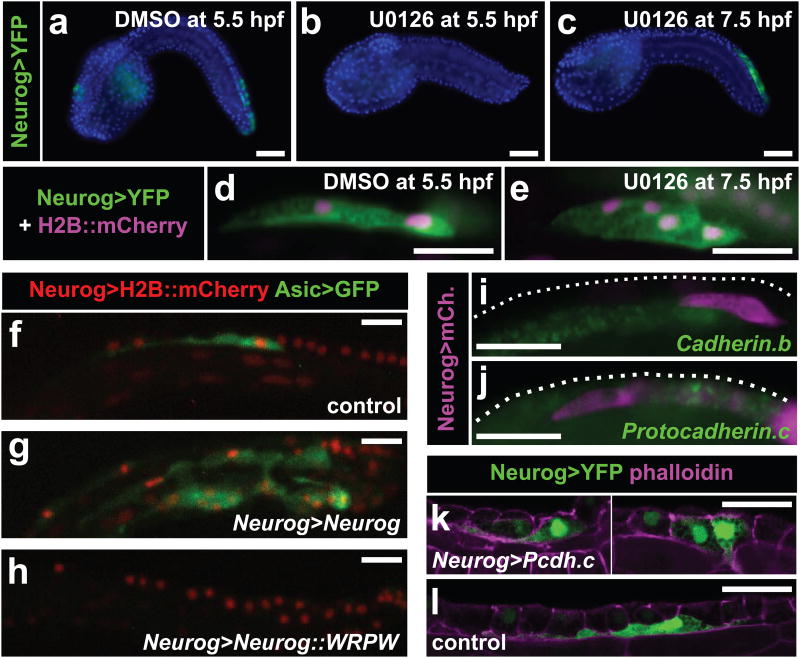
Figure 3. Bipolar tail neuron specification and differentiation.
b, Wild-type Neurogenin (Neurog) b-line>unc-76∷VenusYFP expression (green) in embryos treated with DMSO vehicle, counterstained with DAPI (blue). b, Neurog expression was abolished in 43/50 embryos treated with 10 μM MEK inhibitor U0126 at 5.5 hpf. c, Supernumerary bipolar tail neurons (BTNs) were specified in 28/50 embryos treated with 10 μM U0126 at 7 hpf. d, Two BTN precursors migrating on one side of a DMSO-treated embryo. e, Expanded chain of four BTNs resulting from treatment with U0126 at 7 hpf. f, BTN expressing Asic>unc-76∷eGFP reporter in embryo electroporated with Neurog b-line>nls∷lacZ as a control. g, Overexpression of Neurog induces specification of ectopic Asic+ BTNs in 53/100 embryos. h, Overexpression of a dominant repressor form of Neurog (Neurog∷WRPW) abolishes BTNs in 97/100 embryos. i, In situ hybridization reveals expression of Cadherin.b (green) in the neural tube but not migrating BTN precursors, and j, expression of Protocadherin.c (green) in dorsal epidermis midline but not BTNs. Embryos in i,j electroporated with Neurog b-line>unc-76∷mCherry (immunolabeling of mCherry in magenta). k, Forced overexpression of Protocadherin.c in the BTN lineage inhibits delamination and migration of BTNs in 7/14 embryos. l, Normal BTNs as seen in 9/12 control embryos (overexpression of β-galactosidase instead). Embryos in k,l electroporated with Neurog b-line>unc-76∷VenusYFP and Neurog b-line>H2B∷VenusYFP (green) and counterstained with phalloidin (magenta). All scale bars, 25 μm. Embryos in a-e, i, j fixed at stage 22. Embryos in f-h, k,l at stage 23.
In vertebrates, neural crest EMT is effected in part through differential cell adhesion, mediated by various mechanisms regulating cadherin function21. We found that expression of Cadherin.b, the predominant cadherin gene expressed in the neural tube of Ciona embryos, is absent in BTN precursors (Fig. 3i). Moreover, BTN precursors do not express Protocadherin.c, a cadherin superfamily gene expressed in caudal epidermal sensory neurons (CESNs) and epidermis midline (Fig. 3j). Overexpression of Protocadherin.c protein inhibited delamination and migration of BTNs (Fig. 3k,l), suggesting that Ciona BTNs and vertebrate neural crest share regulatory strategies for EMT via differential cell-cell adhesion.
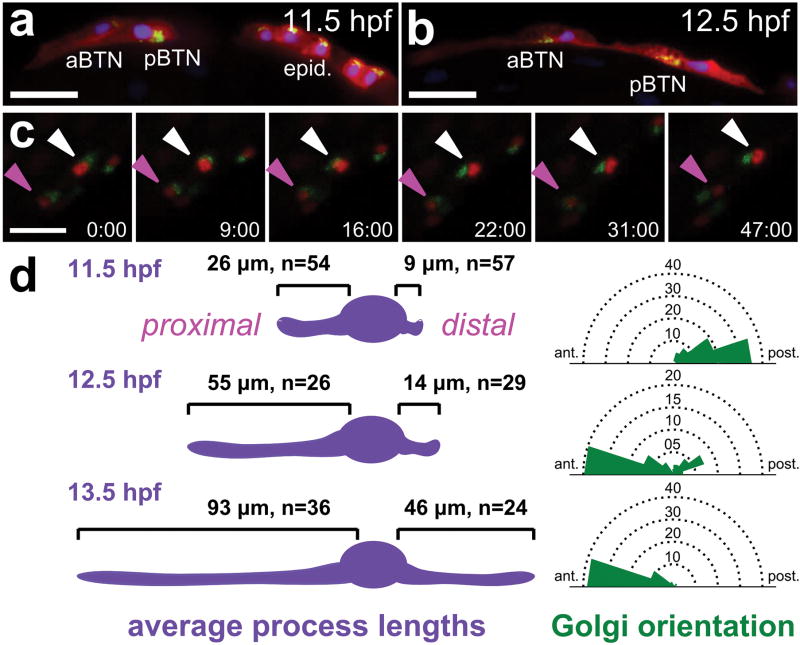
We observed that each BTN precursor initially migrates anteriorly with a prominent leading edge that becomes the cell's anterior neurite (or “proximal process”), while its Golgi apparatus is located posterior to the cell nucleus. At around 12 hours post-fertilization (hpf), each BTN precursor undergoes a 180° polarity inversion, with the Golgi repositioning itself anterior to the nucleus immediately before the cell begins to elaborate the posterior segment of its neurite (the “distal process”), resulting in a bipolar morphology (Extended Data Fig. 6, Supplementary Video 4, and Supplementary Table 1). These observations suggest that a precisely timed re-orientation of cell polarity underlies the characteristic bipolar morphology of the BTNs.
At hatching, BTN cell bodies are situated in the middle of the tail along the anterior-posterior axis, with their distal processes extending towards the tail tip and proximal processes projecting towards the motor ganglion and brain (Fig. 4a-c)12. Electron microscopy confirmed that the BTN somata lie outside the neural tube and are invariably overlain by epidermal cells (Fig. 4d). BTNs lack junctions with epidermal cells and also lack cilia, thus failing to penetrate the tunic to contact the exterior. These characteristics suggest that while distal BTN neurites may be sensory, their cell bodies lack epidermal sensory receptors found in CESNs22. Along the tail, the BTNs contact overlying CESNs, the short processes of which do not reach the motor ganglion12 (Fig. 4a-c). At these contacts synapses form from the CESN to the BTNs (Fig. 4d). Unlike the CESNs, the proximal processes of the BTNs form synaptic contacts with the motor neurons that innervate and control the tail muscles (Fig. 4b,c,e). Each BTN establishes many such contacts upon the two most anterior pairs of motor neurons, MN1 and MN2, on both left and right sides (Fig. 4e, Extended Data Table 1). These synaptic connections are similar to those of mammalian slowly adapting type I DRGNs that, in addition to being mechanosensitive themselves, relay distinct inputs from mechanosensory Merkel cells of the epidermis23. Both tunicate CESNs and vertebrate Merkel cells arise from non-migratory epidermal cells, require Atoh factors for their specification and are glutamatergic in their neurotransmitter phenotype10,20,24,25. These data suggest that tunicate BTNs may thus be equivalent to vertebrate DRGNs within a homologous ascending sensory pathway (Fig. 4c).
Figure 4. Synaptic connections of bipolar tail neurons.
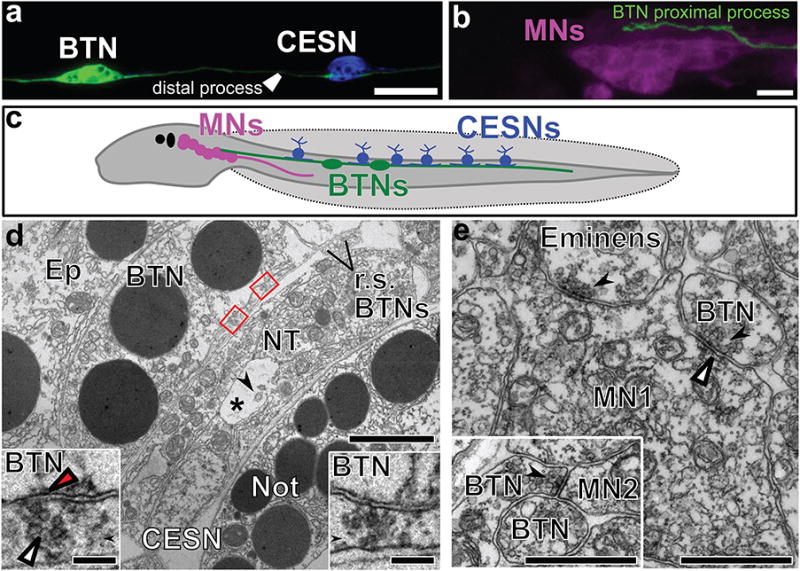
a, Bipolar tail neuron (BTN) labeled by Glutamate decarboxylase>unc-76∷eGFP (green) contacting a caudal epidermal sensory neuron (CESN) labeled by Slc17a6/7/8(Vglut)>unc-76∷mCherry (blue). b, Proximal process of BTN labelled by Islet BTN>unc-76∷mCherry (green) contacting motor ganglion neurons (MNs) labelled by Fgf8/17/18>unc-76∷eGFP (magenta). c, Diagram of Ciona larva showing synaptic connections between CESNs in tail epidermis, BTNs, and MNs. d, Two synaptic inputs (red boxes, insets) from the sheet-like profile of a CESN to a left-side BTN; Transmission electron micrograph from wide-area montage. The profile of a second BTN axon lies out of view. Axon profiles from two right-side BTN axons (r.s. BTNs) are visible. BTNs overlie the neural tube (NT) with neural canal (*) and cross-sectioned cilia (arrow). An epidermal cell (Ep) overlies the BTN. Each synapse enlarged in inset has ∼52 nm diameter presynaptic vesicles (white arrowhead), and the left synapse has a postsynaptic density (red arrowhead). Scale bars 1μm and (insets) 0.5 μm. Not, notochord. e, Synaptic input (arrow) from a BTN to the axon of a member of the most anterior pair (A11.118) of motor neurons (MN1), identified by a cumulus of ∼60-nm presynaptic vesicles and a shallow postsynaptic density (arrowhead). A second input nearby originates from the axon of an Eminens neuron12 (arrow). Inset: Synaptic input from BTN to the axon of a second pair of motor neurons (MN2). Scale bars, 1μm.
In anamniote vertebrates, evidence for a common progenitor of intramedullary Rohon-Beard neurons (RBNs) and neural crest, in addition to other similarities between RBNs and DRGNs, argue for a deep homology between these cell types26. Fritzsch and Northcutt proposed that a key step in the evolution of neural crest was the elaboration of extramedullary sensory neurons from intramedullary RBN-like neurons27. Following the Fritzsch-Northcutt model, the BTNs may be derived from an “intermediate” extramedullary neuron that evolved in the last common ancestor of Olfactores (vertebrates + tunicates) prior to the appearance of bona fide neural crest in the vertebrates. The migration of BTN precursors along the paraxial mesoderm, similar to later phases of DRGN migration, suggests that some of the diverse EMT and migratory behaviors displayed by vertebrate neural crest cells may pre-date the emergence of vertebrates.
Although the embryological origin (neural plate borders) and molecular signature (Neurog+/Islet+) of the BTNs of Ciona also support homology with RBNs, the two do in fact differ in several key aspects. First, BTNs are extramedullary neurons derived from progenitor cells that migrate along paraxial mesoderm lateral to the neural tube. Second, expression of ASICs is shared between BTNs and DRGNs, but appears absent from RBNs28. Finally, RBNs are multipolar with extensively branching peripheral neurites that innervate the overlying epidermis29, while we have not observed any peripheral neurites projecting from the bipolar/pseudounipolar BTNs.
We have revealed the developmental history of migratory neuronal progenitors that arise from the neural plate borders of tunicate embryos. Based on their embryological origin, gene expression, cell behavior, morphology, and synaptic connections, we propose that the BTNs are homologous to neural crest-derived DRGNs. This would imply that the neural plate borders of the olfactorean ancestor gave rise to at least two types of neural crest derivatives: pigment cells and peripheral neurons (Extended Data Fig. 7).
In the invariantly developing Ciona embryo, the pigment cell and BTN lineages become separated early in development, but converge at a neural plate border cell identity before parting again towards distinct differentiated fates. This separation between the two lineages may represent the ancestral condition of the neural plate borders prior to the evolution of the neural crest in vertebrates. This would support models that propose an evolutionary origin for vertebrate neural crest through a heterochronic shift or “intercalation” of a multipotent progenitor state downstream of neural plate border specification but upstream of cell differentiation, based on shared regulatory programs between neural crest and pluripotent cells of the early embryo1,30.
Methods
Molecular cloning
Reporter constructs were designed based on information of cis-regulatory modules (CRMs) from previously published studies on the following genes: Islet31, Msx32, Neurog33, Nodal34, Asic14, Glutamate decarboxylase (Gad)35, Slc17a6/7/8 (Vglut)25, and Fgf8/17/1836. The Neurog b-line CRM (Ciinte.REG.KhC6.1500090-1502346) was cloned using the following primers: Neurog -3010 fwd (5′-gtctgtttccgcatacatgc-3′) and Neurog -773 rev (5′-cttatacgccgaacctcatg-3′). The Neurog b-line minimal CRM (Ciinte.REG.KhC6.1500090-1500501) was found to be contained within this region and cloned using Neurog -3010 fwd and Neurog -2599 rev (5′-gcaaaacgtttcccgattcg-3′) primers. Neurog CRMs were cloned upstream of the basal promoter of Neurog (Ciinte.REG.KhC6.1502506-1503107), cloned using the primers Neurog -594 fwd (5′-ggtcatgctttgttacgtcc-3′) and Neurog +9 rev (5′- atccaacattttgtagcaagagc-3′), or the basal promoter of the Zfpm gene (also known as Friend of GATA, or Fog)37. The full-length Mrf CRM (Ciinte.REG.KhC14.4311719-4314636) was cloned using the primers (5′-gcaagctcctttggggtttgg-3′) and (5′-cgtataaatatgtcaaactaccggc-3′). Caenorhabditis elegans UNC-76 tags were fused to fluorescent proteins to ensure even labeling of axons38. Probes used for in situ hybridization were transcribed in vitro from templates obtained from previously published gene collection clones39,40 for Neurog (R1CiGC29n04), Pax3/7 (R1CiGC42e20), Ebf (R1CiGC02i14), and Cadherin.b (VES104_F13) or cloned de novo from coding sequence for Snail (KH.C3.751.v1.C.SL1-1) and Protocadherin.c (KH.C9.32.v1.A.SL1-1). Golgi-targeting sequence was cloned from KH.C14.396.v1.B.ND1-1 cDNA (N-acetylgalactosaminyltransferase 7, or Galnt7) using the primers Galnt7 a.a.1 fwd (5′-atgagatttaaaatcgcatcagttttg-3′) and Galnt7 a.a.157 rev (5′-aagtgatatcttgtcgctgttcac-3′) and fused in-frame to fluorescent proteins. Neurog coding sequence and Neurog∷WRPW were previously cloned and published41. dnFGFR was previously published42, as was Su(H)-DBM43.
Embryo handling, in situ hybridization and immunolabeling
For purposes other than for electron microscopy (see below) eggs and embryos from wild-caught Ciona intestinalis (species type A, “robusta”) purchased from M-REP (San Diego, USA) were handled according to established protocols44. Double in situ hybridization/immunolabeling was performed as described in previous publications45,46. Monoclonal anti-β-galactosidase (Promega catalogue number Z3781), rabbit polyclonal anti-mCherry (BioVision, accession number ACY24904), and Alexa Fluor-conjugated secondary antibodies (Life Technologies) were all used at 1:500 working dilution. Alexa Fluor-conjugated phalloidin (Life Technologies) was used at 1:50 working dilution. MEK inhibitor U0126 (Cell Signaling Technology) was resuspended as stock solution in DMSO at 10 mM concentration, and diluted to 10 μM in artificial sea water for embryo treatments. Sample sizes equal the total number of embryos present per microscope slide, unless these exceeded arbitrarily set limits of 50 or 100 embryos. No statistics were used. No randomization was used. Replicates were not used. Investigators were not blinded.
Fluorescence/confocal microscopy and photoconversion
Images were captured on a Leica inverted TCS SP8 X confocal or DM2500 epifluorescence microscope. For time-lapse image capture, embryos were imaged as they developed in sea water-filled chambers on coverslip-bottom petri dishes (MatTek). Confocal image stacks were processed in Leica Application Suite or ImageJ. Video annotations were made using Camtasia software (TechSmith). 3D slices and projections were generated using Imaris (Bitplane) or Volocity (PerkinElmer) software. Kaede∷nls47 was photoconverted as previously described48. Neurite lengths and Golgi apparatus positioning were measured using ImageJ. Not all cells, neurites, and/or Golgi were visible in every embryo. Golgi positioning relative to BTN nuclei was measured in degrees of angle formed between a line traced anteriorly from the nucleus and another line traced through the middle of the Golgi complex. Thus, when the Golgi complex is perfectly aligned anterior to the nucleus, the angle is 0°, whereas if the Golgi complex is perfectly posterior to the nucleus, the angle is 180°. Rose plots (angle histograms) were generated in Matlab (http://www.mathworks.com/help/matlab/ref/rose.html).
Electron microscopy
Adult animals, Ciona intestinalis (L.), were collected by Peter Darnell from Mahone Bay, Nova Scotia. Two-hour larvae reared at 18°C in the dark were fixed at 4°C for 1h in 1% OsO4 in 1.25% NaHCO3 adjusted to pH 7.2 with HCl, followed by 2% glutaraldehyde in 0.1M phosphate buffer. After fixation they were embedded in Epon, and a single larva cross sectioned at 60nm in the motor ganglion and later at 100nm down the length of the tail, and the sections post-stained for 5-6 min in freshly prepared aqueous uranyl acetate followed by 2-3 min in lead citrate. Sections were viewed using an FEI Tecnai 12 electron microscope operated at 80kV and images captured using either a Kodak Megaview II camera using software (AnalySIS: SIS GmbH, Münster, Germany), or a Gatan 832 Orius SC1000 CCD camera using Gatan DigitalMicrograph software to compile multi-panel montages from each section. Comprehensive electron micrograph series identified the cell bodies and axons of BTNs, MNs and CESNs from their positions and shapes, and these in turn enabled identification of their connections (K.R. and I.A.M., in preparation).
Extended Data
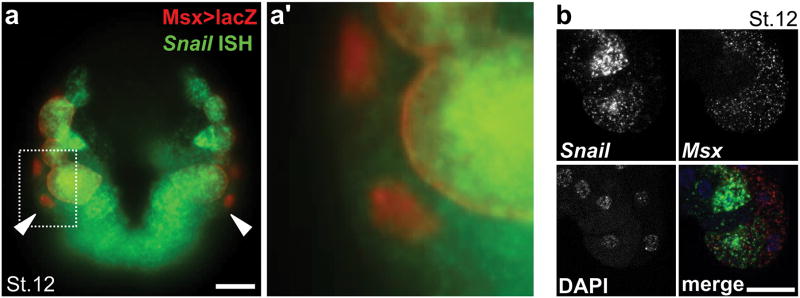
Extended Data Figure 1. In situ hybridization of neural plate border markers Snail and Msx.
a, Immunolabeling for β-Galactosidase (red) and in situ hybridization (ISH) for Snail mRNA (green) in stage 12 embryo electroporated with Msx>lacZ, revealing Snail expression in the bipolar tail neuron (BTN) progenitors (b9.36 cells, arrowheads). Dashed area enlarged in a′. b, Double ISH for Snail and Msx in stage 12 embryos counterstained with DAPI, showing co-expression in neural plate border cells, including BTN progenitors. Scale bars, 25 μm.
Extended Data Figure 2. Lineage tracing of b9.36 descendants.
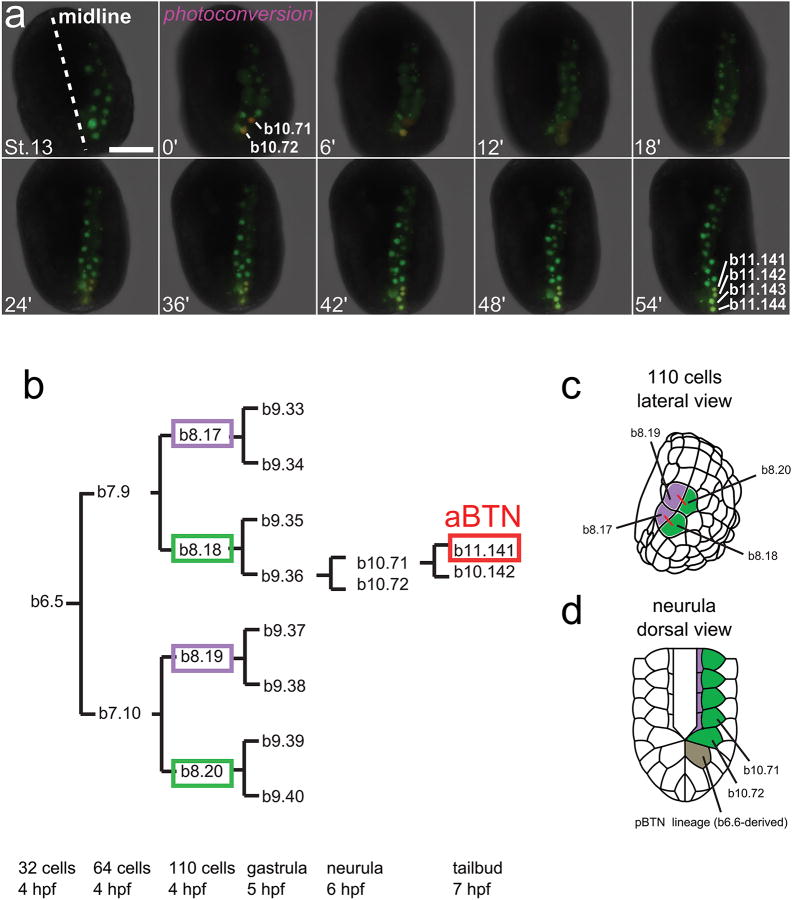
a, Photoconversion of Kaede∷nls driven by the Msx driver was used to follow the cell divisions of the bipolar tail neuron (BTN) progenitors from the late gastrula stage to the early tailbud stage. Both b10.71 and b10.72 divide once. b11.141 will later divide again to give rise to a definitive anterior BTN (see Extended Data Figure 4). Numbers in each panel represent time in minutes elapsed from initial photoconversion event. Scale bar, 50 μm. b, Lineage tree showing specification of anterior BTNs (aBTN) in relation to other cells of the posterior neural plate borders. For simplicity, only one side of the embryo is depicted. c, Lateral view of a 110-cell-stage embryo showing the positions of blastomeres in b. Red lines connect sibling cells. d, Dorsal view of a neurula-stage embryo showing zippering of posterior neural plate border-derived capstone cells18 as neural tube closure is initiated. Panels b, d are courtesy of Dr. Hidehiko Hashimoto and Dr. François Robin (University of Chicago) and Dr. Naohito Takatori (Tokyo Metropolitan University), and partially modeled after Hashimoto et al. 201517. Panel c modeled after Nishida 198649.
Extended Data Figure 3. Neurogenin cis-regulatory sequences.
a, Schematic diagram representing Neurogenin (Neurog) locus and 5′ cis-regulatory sequences including “b-line” and “b-line minimal” cis-regulatory modules. Peaks represent nucleotide sequence conservation with Ciona savignyi genome. b, Late gastrula embryo (stage 13) electroporated with full length Neurog (blue) and Nodal b-line (red) reporter constructs. Reporter co-expression is seen in b9.36 descendants on either side of the neural plate. Neurog expression also marks tail tip lineages of uncertain provenance, previously reported to be descended from b8.2110. Scale bar, 25 μm. c, Neurog b-line reporter. d, Neurog b-line minimal reporter. Scale bars in c, d 50 μm.
Extended Data Figure 4. Spatiotemporal restriction of Neurogenin expression.
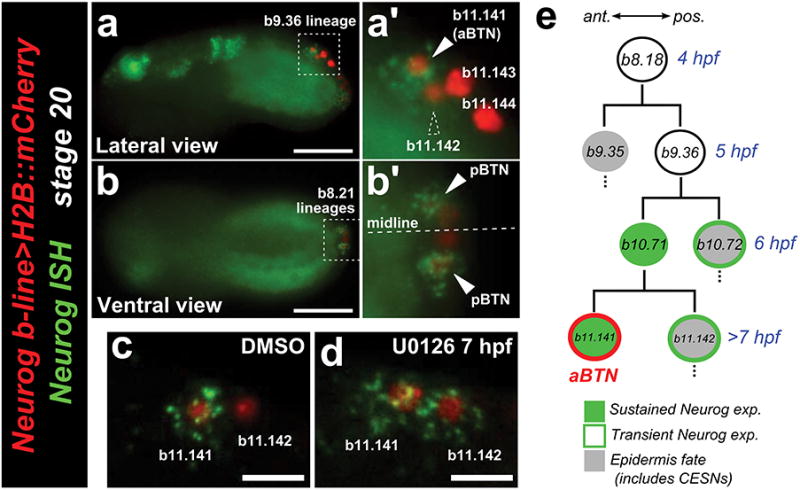
a, Lateral view of in situ hybridization (ISH) for Neurogenin (Neurog, green) in embryo electroporated with Neurog b-line>H2B∷mCherry (red) shows that Neurog expression is selectively maintained in only a subset of initially Neurog-expressing neural plate border cells. a′, In the b9.36 lineage, the anterior-most cell (b11.141, solid arrowhead) is always the sole one to express Neurog at this stage, and will go on to become the anterior bipolar tail neuron (aBTN). Dashed arrowhead indicates b11.142, the sister cell of b11.141, which has downregulated Neurog relative to its sibling. b, b′, The identities of the cells in the tail tip (presumed b8.21-derived) lineages are unclear, but Neurog is similarly restricted (arrowheads) to a single cell on either side of the midline, which we interpret as the definitive posterior BTNs (pBTNs). c, Control embryo treated with DMSO vehicle, showing wild-type pattern of Neurog expression only in b11.141. d, Neurog is expanded to b11.142 upon treatment with the MEK inhibitor U0126 at 7 hpf. This condition also results in specification of supernumerary BTNs, presumably due to expanded Neurog expression (see text for details). Thus, downregulation of Neurog in b11.142 also requires MEK/ERK signaling. e, Diagram of the aBTN lineage, descended from the b8.18 blastomere. Scale bars in a,b 25 μm. Scale bars in c,d 10 μm.
Extended Data Figure 5. Perturbation of Notch signaling does not alter Neurogenin expression or bipolar tail neuron specification and differentiation.
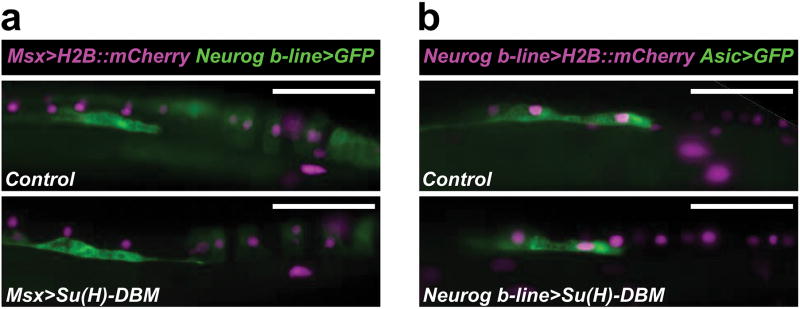
a, Upper panel, lateral view of a stage 23 embryo electroporated with Msx>H2B∷mCherry (magenta nuclei) and Neurogenin (Neurog) b-line>unc-76∷eGFP (green) and Msx>nls∷lacZ, serving as the wild-type “control” condition. Lower panel, embryo electroporated with same reporters as upper panel, plus Msx>Su(H)-DBM, which encodes a DNA-binding mutant form of the Notch co-activator Rbpj. No discernable difference in Neurog activation or bipolar tail neuron (BTN) specification was observed between control and Su(H)-DBM conditions (1/32 vs. 2/42 embryos showing ectopic Neurog+ BTNs, respectively). b, Late overexpression of Su(H)-DBM using the Neurog b-line driver similarly did not alter BTN specification/differentiation, as monitored by Asic>unc-76∷eGFP reporter expression (0/50 control vs. 0/50 Su(H)-DBM embryos showed ectopic Asic+ BTNs). All scale bars 50 μm.
Extended Data Figure 6. Cell polarity and morphogenesis of bipolar tail neurons.
a, Embryo at 11.5 hpf (18°C) with bipolar tail neurons (BTNs) displaced from clonally related epidermal cells (“epid.”) labeled by UNC-76∷VenusYFP (red), Galnt7ΔC∷CFP (green), and H2B∷mCherry (blue) driven by Neurogenin b-line cis-regulatory module. Targeted localization of CFP by the Galnt7 N-terminal signal sequence reveals polarized subcellular distribution of Golgi apparatus on posterior side of BTN nuclei as migration and proximal process extend in an anterior direction. This is distinct from the apical (dorsal) location of the Golgi apparatus in epidermal cells. b, Embryo at 12.5 hpf (18°C) showing 180° inversion of Golgi apparatus localization to the anterior side of the nucleus, immediately preceding distal process extension. Scale bars in a,b 50 μm. c, Still frames from a confocal image stack time lapse movie (Supplementary Video 4) showing inversion of Golgi complex (Galnt7ΔC∷VenusYFP, green) relative to nuclei (H2B∷mCherry, red) in migrating BTNs. Time lapse imaging initiated at 11.5 hpf (18°C). Time in minutes elapsed from start shown at bottom right of each panel. Anterior BTN (aBTN) indicated by magenta arrowhead, posterior BTN (pBTN) indicated by white arrowhead. Scale bar, 25 μm. d, Diagram showing correlation of average length of proximal (left) and distal (right) processes and angle of Golgi apparatus location relative to cell nucleus along the anterior-posterior axis in BTNs at different time points. Locations of Golgi apparatus represented by rose plots of bins of 20° spanning anterior (0°) and posterior (180°) endpoints around dorsal edge of BTN nucleus. Bin diameters indicate number of cells. Embryos analyzed belong to the same pool as embryos in a,b. See Supplementary Table 1 for source data.
Extended Data Figure 7. Proposed evolution of Neural Crest through the acquisition of multipotency by neural plate border cells.
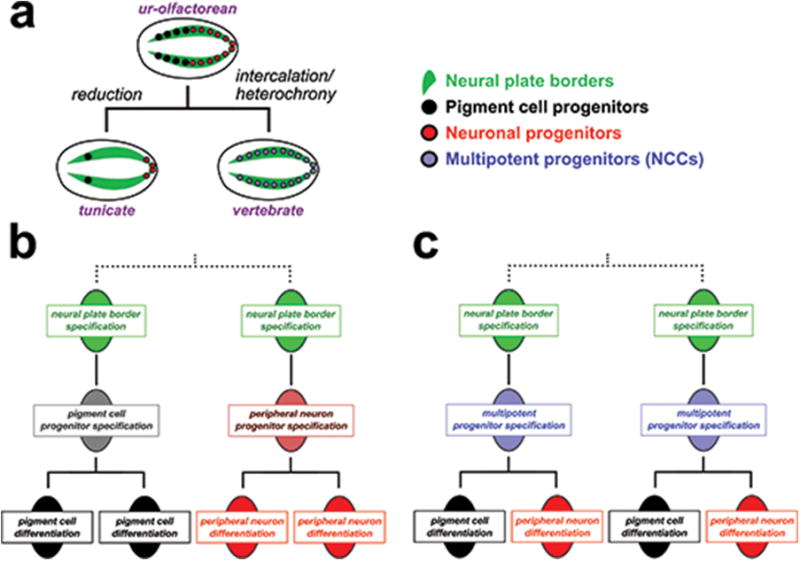
a, Cartoon diagram depicting a hypothetical path for neural plate border and neural crest evolution, starting with the reconstructed ur-olfactorean ancestor, which could have had neural plate borders lined with committed progenitor cells giving rise to several pigmented ocelli and bipolar tail neuron (BTN)-like peripheral neurons, a condition that may be conserved in extant cephalochordates50. These cells would have been reduced in the highly miniaturized embryos of extant tunicates, while vertebrates are proposed to have co-opted a mesenchymal, multipotency program to bestow these cells with the potential to give rise to pigment cells, peripheral neurons, or other derivatives, after a prolonged period of epithelial-to-mesenchymal transition and migration. b, Diagram representing idealized cell lineages in the neural plate borders of tunicate and hypothetical ur-olfactorean ancestor, in which segregated lineages at the neural plate borders give rise to committed pigment cell or peripheral neuronal progenitors. c, Diagram of simplified neural crest cell lineage deploying a multipotency program downstream of neural plate border specification and upstream of cell differentiation. Thus, neural crest cells could have evolved through redeployment of a multipotency program (“intercalation” hypothesis)1, or through its maintenance from earlier embryonic stages (“heterochrony” hypothesis)30.
Table Extended Data 1. Synaptic input from bipolar tail neurons to motor neurons, identified by electron microscopy.
| Postsynaptic motor neuron identity | Synapse partnership | Number of synapses | Total number of sections with synaptic profile |
|---|---|---|---|
| MN1 Left (A11.118) | BTN1-->MN1L | 27 | 134 |
| BTN3-->MN1L | 21 | 88 | |
| Total | 48 | 222 | |
|
|
|||
| MN1 Right (A11.118) | BTN1-->MN1R | 3 | 14 |
| BTN2-->MN1R | 22 | 94 | |
| BTN3-->MN1R | 1 | 4 | |
| BTN4-->MN1R | 11 | 55 | |
| Total | 37 | 167 | |
|
|
|||
| MN2 Left (A10.57) | BTN1-->MN2L | 10 | 51 |
| BTN3-->MN2L | 6 | 30 | |
| Total | 16 | 81 | |
|
|
|||
| MN2 Right (A10.57) | BTN2-->MN2R | 17 | 90 |
| BTN4-->MN2R | 10 | 73 | |
| Total | 27 | 163 | |
|
|
|||
| MN3 Left | BTN1-->MN3L | 1 | 2 |
| Total | 1 | 2 | |
|
|
|||
| MN4 Left | BTN1-->MN4L | 2 | 9 |
| Total | 2 | 9 | |
|
|
|||
| MN4 Right | BTN2-->MN4R | 2 | 5 |
| BTN4-->MN4R | 1 | 2 | |
| Total | 3 | 7 | |
|
|
|||
| MN5 Left | BTN1-->MN5L | 1 | 3 |
| Total | 1 | 3 | |
|
|
|||
| MN5 Right | BTN4-->MN5R | 1 | 3 |
| Total | 1 | 3 | |
BTN, bipolar tail neuron. MN, motor neuron. Axons of BTN 1 and 3 lie on the left hand side of the embryo, and BTN 2 and 4 on the right. The axons are not traced to their somata to indicate which would be anterior and posterior.
Supplementary Material
Acknowledgments
The authors would like to thank Florian Razy-Krajka for assistance with Kaede photoconversion and comments on the manuscript, Theadora Tolkin for constructing the Mrf reporter plasmid, Zhiyuan Lu for ultramicrotomy, and Claude Desplan, Anna Di Gregorio and all members of the Christiaen and Meinertzhagen labs for feedback and suggestions. We thank Hidehiko Hashimoto, François Robin, and Naohito Takatori for embryo illustration template files. This work was funded by a National Science Foundation Postdoctoral Fellowship in Biology (under grant NSF-1161835) to A.S., by National Institutes of Health award GM096032 to L.C., and by grant DIS0000065 from NSERC (Ottawa) to I.A.M.
Footnotes
The Authors declare no competing financial interests.
A.S., K.R., I.A.M., and L.C. designed the study, analyzed the data, and wrote the paper. A.S. and K.R. performed the experiments.
References
- 1.Bronner ME, LeDouarin NM. Evolution and Development of the Neural Crest: An Overview. Developmental Biology. 2012;366:2–9. doi: 10.1016/j.ydbio.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu JK, Meulemans D, McKeown SJ, Bronner-Fraser M. Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Research. 2008;18:1127–1132. doi: 10.1101/gr.076208.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wada H, Holland PWH, Sato S, Yamamoto H, Satoh N. Neural Tube Is Partially Dorsalized by Overexpression of HrPax-37: The Ascidian Homologue of Pax-3and Pax-7. Developmental Biology. 1997;187:240–252. doi: 10.1006/dbio.1997.8626. doi: http://dx.doi.org/10.1006/dbio.1997.8626. [DOI] [PubMed] [Google Scholar]
- 4.Abitua PB, Wagner E, Navarrete IA, Levine M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature. 2012;492:104–107. doi: 10.1038/nature11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wada H. Origin and evolution of the neural crest: a hypothetical reconstruction of its evolutionary history. Development, growth & differentiation. 2001;43:509–520. doi: 10.1046/j.1440-169x.2001.00600.x. [DOI] [PubMed] [Google Scholar]
- 6.Baker CVH, Bronner-Fraser M. The origins of the neural crest. Part II: an evolutionary perspective. Mechanisms of Development. 1997;69:13–29. doi: 10.1016/s0925-4773(97)00129-9. doi: http://dx.doi.org/10.1016/S0925-4773(97)00129-9. [DOI] [PubMed] [Google Scholar]
- 7.Shimeld SM, Holland PWH. Vertebrate innovations. Proceedings of the National Academy of Sciences. 2000;97:4449–4452. doi: 10.1073/pnas.97.9.4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeffery WR, Strickler AG, Yamamoto Y. Migratory neural crest-like cells form body pigmentation in a urochordate embryo. Nature. 2004;431:696–699. doi: 10.1038/nature02975. [DOI] [PubMed] [Google Scholar]
- 9.Mazet F, et al. Molecular evidence from Ciona intestinalis for the evolutionary origin of vertebrate sensory placodes. Developmental biology. 2005;282:494–508. doi: 10.1016/j.ydbio.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Pasini A, et al. Formation of the ascidian epidermal sensory neurons: insights into the origin of the chordate peripheral nervous system. PLoS Biology. 2006;4:e225. doi: 10.1371/journal.pbio.0040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaltenbach SL, Yu JK, Holland ND. The origin and migration of the earliest-developing sensory neurons in the peripheral nervous system of amphioxus. Evolution & Development. 2009;11:142–151. doi: 10.1111/j.1525-142X.2009.00315.x. [DOI] [PubMed] [Google Scholar]
- 12.Imai JH, Meinertzhagen IA. Neurons of the ascidian larval nervous system in Ciona intestinalis: II. Peripheral nervous system. The Journal of comparative neurology. 2007;501:335–352. doi: 10.1002/cne.21247. [DOI] [PubMed] [Google Scholar]
- 13.Ma Q, Fode C, Guillemot F, Anderson DJ. NEUROGENIN1 and NEUROGENIN2 control two distinct waves of neurogenesis in developing dorsal root ganglia. Genes & Development. 1999;13:1717–1728. doi: 10.1101/gad.13.13.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coric T, Passamaneck YJ, Zhang P, Di Gregorio A, Canessa CM. Simple chordates exhibit a proton-independent function of acid-sensing ion channels. The FASEB Journal. 2008;22:1914–1923. doi: 10.1096/fj.07-100313. [DOI] [PubMed] [Google Scholar]
- 15.Aniello F. et al. Identification and developmental expression of Ci-msxb: a novel homologue of Drosophila msh gene in Ciona intestinalis. Mechanisms of Development. 1999;88:123–126. doi: 10.1016/s0925-4773(99)00178-1. doi: http://dx.doi.org/10.1016/S0925-4773(99)00178-1. [DOI] [PubMed] [Google Scholar]
- 16.Wada S, Saiga H. Cloning and embryonic expression of Hrsna, a snail family gene of the ascidian Halocynthia roretzi: Implication in the origins of mechanisms for mesoderm specification and body axis formation in chordates. Development, Growth & Differentiation. 1999;41:9–18. doi: 10.1046/j.1440-169x.1999.00408.x. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto H, Robin FB, Sherrard KM, Munro EM. Sequential Contraction and Exchange of Apical Junctions Drives Zippering and Neural Tube Closure in a Simple Chordate. Developmental cell. 2015;32:241–255. doi: 10.1016/j.devcel.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Nicol D, Meinertzhagen I. Development of the central nervous system of the larva of the ascidian, Ciona intestinalis L: II. Neural plate morphogenesis and cell lineages during neurulation. Developmental biology. 1988;130:737–766. doi: 10.1016/0012-1606(88)90364-8. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura MJ, Terai J, Okubo R, Hotta K, Oka K. Three-dimensional anatomy of the Ciona intestinalis tailbud embryo at single-cell resolution. Developmental Biology. 2012;372:274–284. doi: 10.1016/j.ydbio.2012.09.007. doi: http://dx.doi.org/10.1016/j.ydbio.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Tang WJ, Chen JS, Zeller RW. Transcriptional regulation of the peripheral nervous system in Ciona intestinalis. Developmental biology. 2013;378:183–193. doi: 10.1016/j.ydbio.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 21.Theveneau E, Mayor R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Developmental Biology. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. doi: http://dx.doi.org/10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 22.Torrence S, Cloney R. Nervous system of ascidian larvae: Caudal primary sensory neurons. Zoomorphology. 1982;99:103–115. doi: 10.1007/BF00310303. [DOI] [Google Scholar]
- 23.Maksimovic S, et al. Epidermal Merkel cells are mechanosensory cells that tune mammalian touch receptors. Nature. 2014;509:617–621. doi: 10.1038/nature13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison KM, Miesegaes GR, Lumpkin EA, Maricich SM. Mammalian Merkel cells are descended from the epidermal lineage. Developmental Biology. 2009;336:76–83. doi: 10.1016/j.ydbio.2009.09.032. doi: http://dx.doi.org/10.1016/j.ydbio.2009.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horie T, Kusakabe T, Tsuda M. Glutamatergic networks in the Ciona intestinalis larva. The Journal of Comparative Neurology. 2008;508:249–263. doi: 10.1002/cne.21678. [DOI] [PubMed] [Google Scholar]
- 26.Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126:3969–3979. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fritzsch B, Northcutt RG. Cranial and spinal nerve organization in amphioxus and lampreys: evidence for an ancestral craniate pattern. Cells Tissues Organs. 1993;148:96–109. doi: 10.1159/000147529. [DOI] [PubMed] [Google Scholar]
- 28.Paukert M, et al. A Family of Acid-sensing Ion Channels from the Zebrafish: Widespread Expression in the Central Nervous System Suggests a Conserved Role in Neuronal Communication. Journal of Biological Chemistry. 1878;2004;279:3–18791. doi: 10.1074/jbc.M401477200. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien GS, et al. Coordinate development of skin cells and cutaneous sensory axons in zebrafish. Journal of Comparative Neurology. 2012;520:816–831. doi: 10.1002/cne.22791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buitrago-Delgado E, Nordin K, Rao A, Geary L, LaBonne C. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science. 2015;348:1332–1335. doi: 10.1126/science.aaa3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolfi A, et al. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo MT, et al. Regulatory elements controlling Ci-msxb tissue-specific expression during Ciona intestinalis embryonic development. Developmental Biology. 2004;267:517–528. doi: 10.1016/j.ydbio.2003.11.005. doi: http://dx.doi.org/10.1016/j.ydbio.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 33.Stolfi A, Christiaen L. Genetic and genomic toolbox of the chordate Ciona intestinalis. Genetics. 2012;192:55–66. doi: 10.1534/genetics.112.140590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khoueiry P, et al. A cis-Regulatory Signature in Ascidians and Flies, Independent of Transcription Factor Binding Sites. Current Biology. 2010;20:792–802. doi: 10.1016/j.cub.2010.03.063. doi: http://dx.doi.org/10.1016/j.cub.2010.03.063. [DOI] [PubMed] [Google Scholar]
- 35.Takamura K, Minamida N, Okabe S. Neural Map of the Larval Central Nervous System in the Ascidian Ciona intestinalis. Zoological Science. 2010;27:191–203. doi: 10.2108/zsj.27.191. [DOI] [PubMed] [Google Scholar]
- 36.Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136:285. doi: 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- 37.Rothbächer U, Bertrand V, Lamy C, Lemaire P. A combinatorial code of maternal GATA, Ets and β-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development. 2007;134:4023–4032. doi: 10.1242/dev.010850. [DOI] [PubMed] [Google Scholar]
- 38.Dynes JL, Ngai J. Pathfinding of Olfactory Neuron Axons to Stereotyped Glomerular Targets Revealed by Dynamic Imaging in Living Zebrafish Embryos. Neuron. 1998;20:1081–1091. doi: 10.1016/s0896-6273(00)80490-0. doi: http://dx.doi.org/10.1016/S0896-6273(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 39.Satou Y, et al. A cDNA resource from the basal chordate Ciona intestinalis. genesis. 2002;33:153–154. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- 40.Roure A, et al. A multicassette Gateway vector set for high throughput and comparative analyses in Ciona and vertebrate embryos. PLoS One. 2007;2:e916. doi: 10.1371/journal.pone.0000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolfi A, Wagner E, Taliaferro JM, Chou S, Levine M. Neural tube patterning by Ephrin, FGF and Notch signaling relays. Development. 2011;138:5429–5439. doi: 10.1242/dev.072108. [DOI] [PubMed] [Google Scholar]
- 42.Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes & development. 2006;20:2728. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hudson C, Yasuo H. A signalling relay involving Nodal and Delta ligands acts during secondary notochord induction in Ciona embryos. Development. 2006;133:2855–2864. doi: 10.1242/dev.02466. [DOI] [PubMed] [Google Scholar]
- 44.Christiaen L, Wagner E, Shi W, Levine M. The sea squirt Ciona intestinalis. Cold Spring Harbor protocols. 2009;2009 doi: 10.1101/pdb.emo138. pdb. emo138. [DOI] [PubMed] [Google Scholar]
- 45.Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- 46.Ikuta T, Saiga H. Dynamic change in the expression of developmental genes in the ascidian central nervous system: revisit to the tripartite model and the origin of the midbrain–hindbrain boundary region. Developmental biology. 2007;312:631–643. doi: 10.1016/j.ydbio.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proceedings of the National Academy of Sciences. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Razy-Krajka F, et al. Collier/OLF/EBF-Dependent Transcriptional Dynamics Control Pharyngeal Muscle Specification from Primed Cardiopharyngeal Progenitors. Developmental cell. 2014;29:263–276. doi: 10.1016/j.devcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishida H. Cell division pattern during gastrulation of the ascidian, Halocynthia roretzi. Dev Growth Differ. 1986;28:191–201. doi: 10.1111/j.1440-169X.1986.00191.x. [DOI] [PubMed] [Google Scholar]
- 50.Bone Q. The central nervous system in amphioxus. Journal of Comparative Neurology. 1960;115:27–64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.