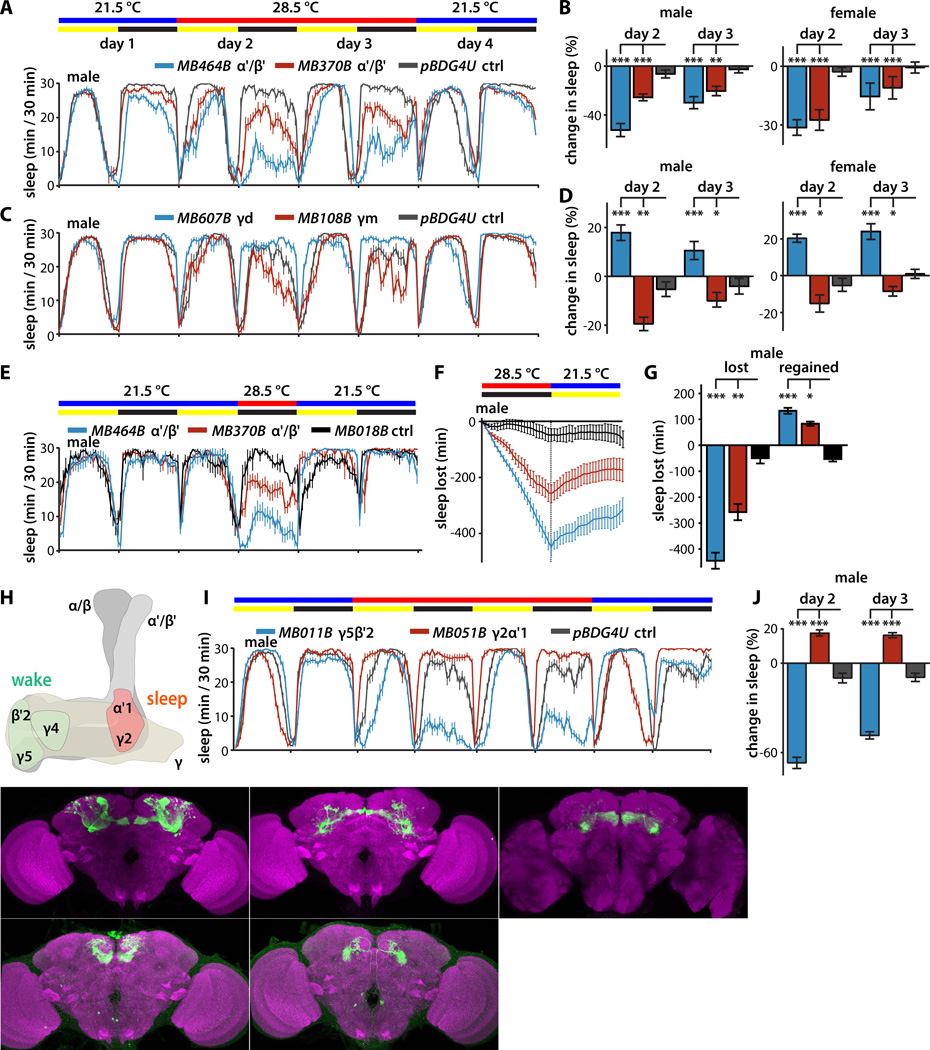
Figure 2. Sleep profiles of flies following thermogenetic activation of KCs and MBONs.
(A) Sleep profiles of male flies expressing dTRPA1 in α'/β' KCs driven by the indicated split-GAL4 lines (mean±sem). Flies were maintained in 12 hr:12 hr light:dark (LD) conditions, and the temperature was increased for days 2 and 3 of the four day experiment.
(B) Quantification of change in sleep relative to day 1 of male and female flies expressing dTRPA1 in α'/β' KCs. Genotypes are color coded as in (A). Statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test.
(C) Sleep profiles of male flies expressing dTRPA1 in γd or γm KCs under the control of the indicated split-GAL4 lines, treated as in (A).
(D) Quantification of change in sleep of male and female flies expressing dTRPA1 in γd or γm KCs. Genotypes are color coded as in (C); statistical analysis as in (B).
(E) Sleep profiles of male flies expressing dTRPA1 in α'/β' KCs driven by the indicated split-GAL4 lines (mean±sem). Flies were maintained in 12 hr:12 hr light:dark (LD) conditions, and the temperature was increased for 12 hr during the night of day 2 of this three day experiment.
(F) Cumulative sleep lost during thermogenetic sleep suppression at 28.5°C and regained during subsequent recovery for 12 hours at 21.5°C (mean±sem). Genotypes are color coded as in (E).
(G) Quantification of sleep lost during thermogenetic sleep suppression and regained during the 12 hours of recovery. Sleep suppression induced by dTRPA1-mediated activation of α'/β' KCs using either of two split-GAL4 drivers resulted in homeostatic rebound compared to control flies expressing dTRPA1 in a cholinergic MBON that does not influence sleep (MB018B). Genotypes are color coded as in (D); statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test. (*, p<0.05; **, p<0.01; ***p<0.001; n=22–24 flies per genotype.)
(H) Schematic representation of the MB lobes indicating the compartment-specific dendritic projections of sleep-regulating MBON-γ5β′2a/β′2mp/β′2mp_bilateral, MBON-γ4>γ1γ2, and MBON-γ2α'1.
(I) Sleep profiles of male flies expressing dTRPA1 in MBONs under the control of the indicated split-GAL4 lines, treated as in (A).
(J) Quantification of change in sleep of male flies expressing dTRPA1 in MBONs. Genotypes color coded as in (F); statistical analysis as in (B).
(K) Whole brains of flies expressing GFP under the control of the indicated MBON and KC split-GAL4 driver lines, immunostained for GFP (green) and the synaptic neuropil marker BRUCHPILOT (magenta).