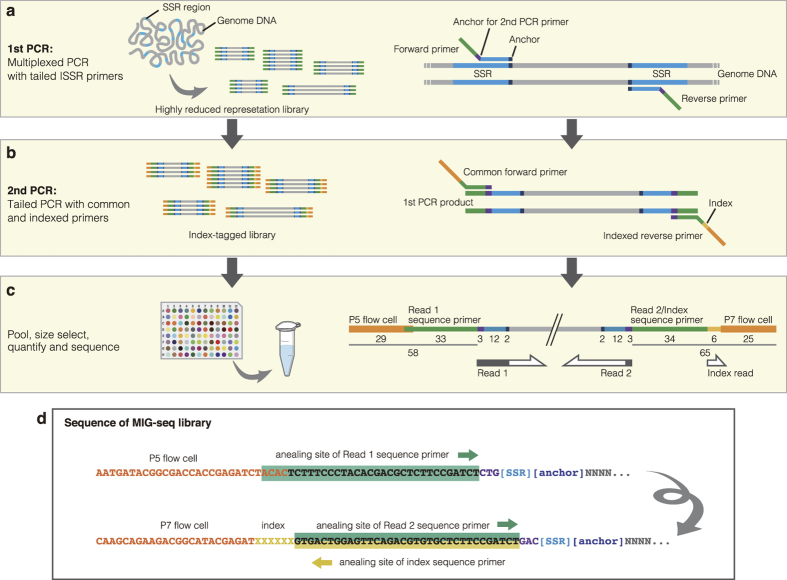
Figure 1. Construction of the MIG-seq library.
(a) Multiple non-repetitive regions from various inter-simple sequence repeats (ISSRs) are amplified from genomic DNA by multiplexed PCR with tailed ISSR primers (1st PCR). (b) The 1st PCR products are subsequently used as the templates for the 2nd PCR (tailed PCR). This step enables the addition of complementary sequences for the binding sites of Illumina sequencing flow cell and indices (barcodes) for each sample to the 1st PCR products, using common forward and indexed reverse primers. (c) After measuring the approximate concentration of each 2nd PCR product, they are pooled in equimolar concentrations as a single mixture library. The mixture is then purified, fragments with a size range of 300–800 bp are isolated, the final concentration is measured by quantitative PCR, and is then used for Illumina paired-end sequencing (reads 1 and 2) and index reading. Sequencing of the first 17 nucleotides (primer region) of read 1, and 3 nucleotides (anchor region) of read 2 are skipped using the ‘DarkCycle’ option of the sequencer (indicated as the gray region in the arrows). (d) The sequence of the resulting library consists of binding sites for the P5 flow cell oligonucleotides and read 1 sequencing primer, forward ISSR primer, DNA insert, reverse ISSR primer, binding sites for read 2 and index sequencing primers, and P7 flow cell oligonucleotides.