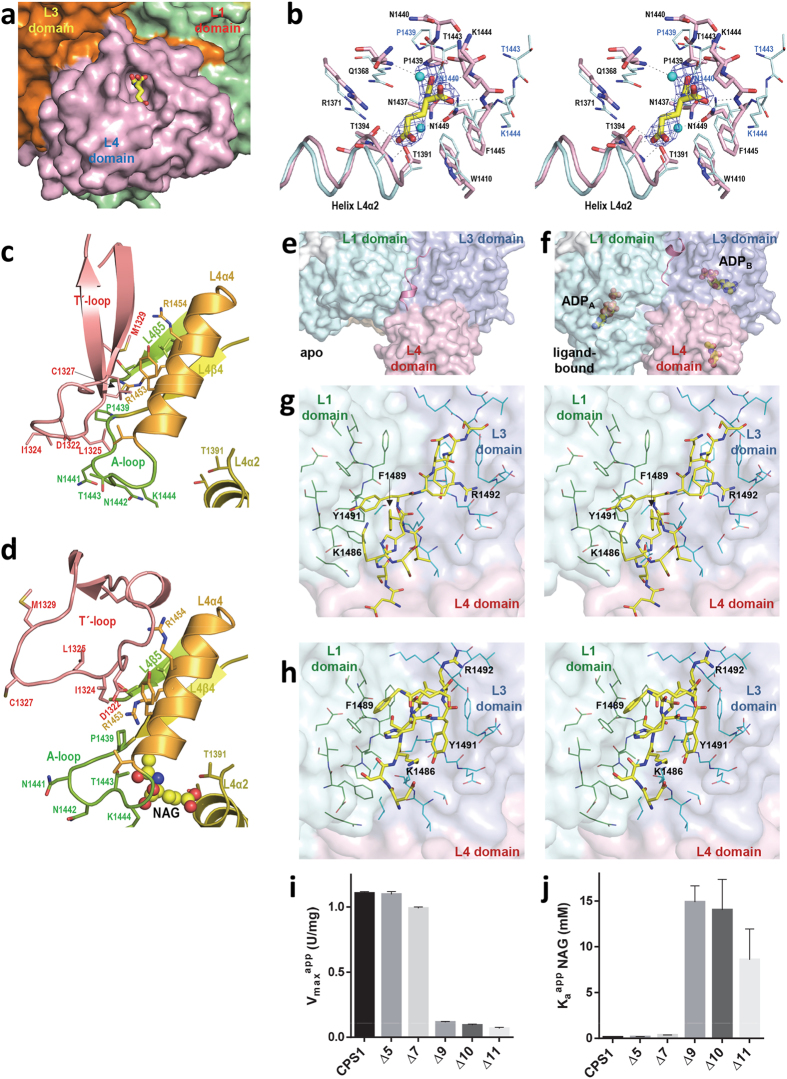
Figure 2. The allosteric L4 domain.
(a) View of NAG bound in its binding pocket in the L4 domain, not far from the interface with the L3 domain. (b) Stereo view of the superposition of the NAG site of the apo (carbon atoms colored cyan) and ligand-bound (protein and NAG carbon atoms colored pink and yellow, respectively) forms, identifying the residues by labeling them. When the position of one residue changes much, the residue is labeled in blue for the apo form. The unbiased (Fo-Fc) omit electron density map obtained prior to building the ligand is shown as a blue grid contoured at 2σ. (c,d) Helix L4α4 and some adjacent regions of the L4 domain and the T′-loop from domain L3 are shown, in the (c) apo and (d) ligand-bound forms. NAG is shown in spheres representation. The side-chains of some residues are shown in sticks representation and some secondary structure elements and residues are identified by labeling in the same color as the structural element to which they belong. (e,f) C-tail from domain L4 interacting with neighbor domains L1 and L3, in the apo and ligand-bound forms, and (g,h) respective close-up stereo-views of (e,f) with some residues labeled to highlight the radical changes of both the C-tail structure and their interactions with neighbor domains. (i,j) Influence of C-terminal truncations of the indicated length (Δn denotes the number of deleted residues starting from the C-terminus) on (g) the activity of the enzyme at saturation of NAG (Vmaxapp), and on (h) the concentration of NAG (KaappNAG) needed for attaining an activity of 0.5 × Vmaxapp. CPS1 denotes the wild-type enzyme.