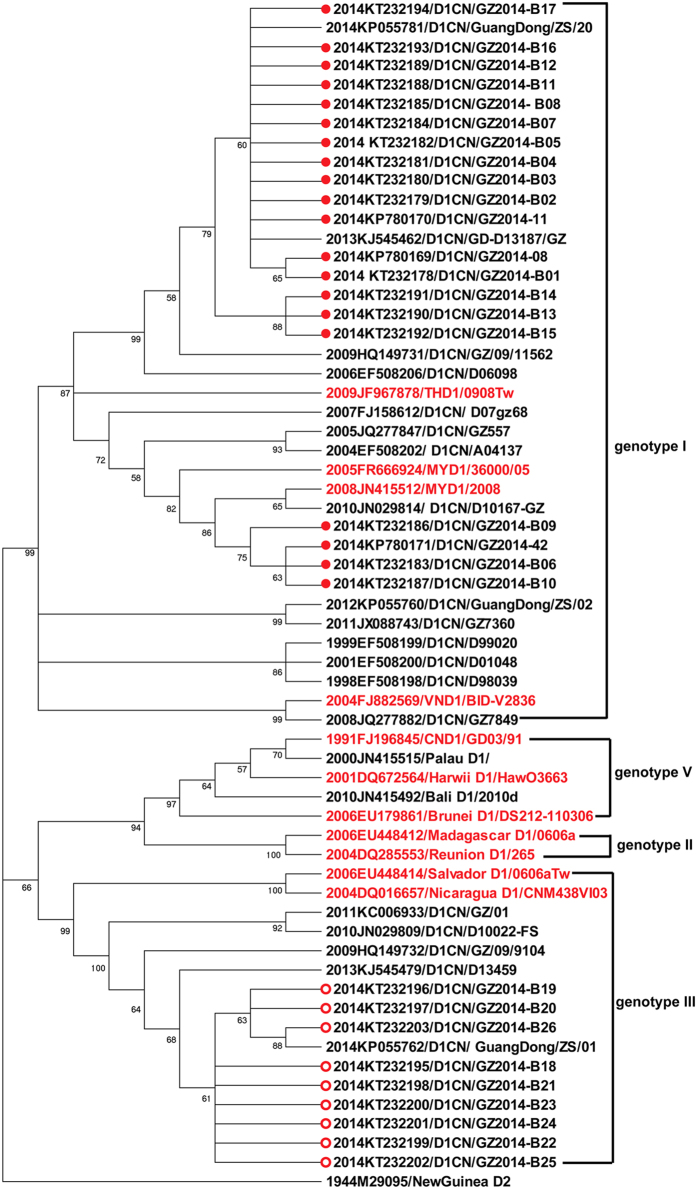
Figure 3. Phylogenetic analysis of isolated DENV-1 along with the homologous strains reported in Guangdong Province.
A phylogenetic tree was constructed by the Neighbor-joining method based on the Tajima-Nei model using MEGA5.21 software. The bootstrap value was set for 1000 repetitions. Branches corresponding to partitions reproduced in fewer than 50% of the bootstrap replicates were collapsed. The percentages of replicate trees in which the associated dengue virus isolates clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The D2 strain of New Guinea reported in 1944 (M29095) was used as an out-group to root the tree. The sequences in red are references for different genotypes (DENV-1 Genotype I: JN415512, FR666924, FJ882569 and JF967878; DENV-1 Genotype V: DQ672564, FJ196845, and EU179861; DENV-1 Genotype III: EU448414 and DQ016657; DENV-1 Genotype II: EU448412 and DQ285553). The strains isolated in this outbreak that were classified as Genotype I are labeled with red dots, and those classified as genotype II are labeled with red circles.