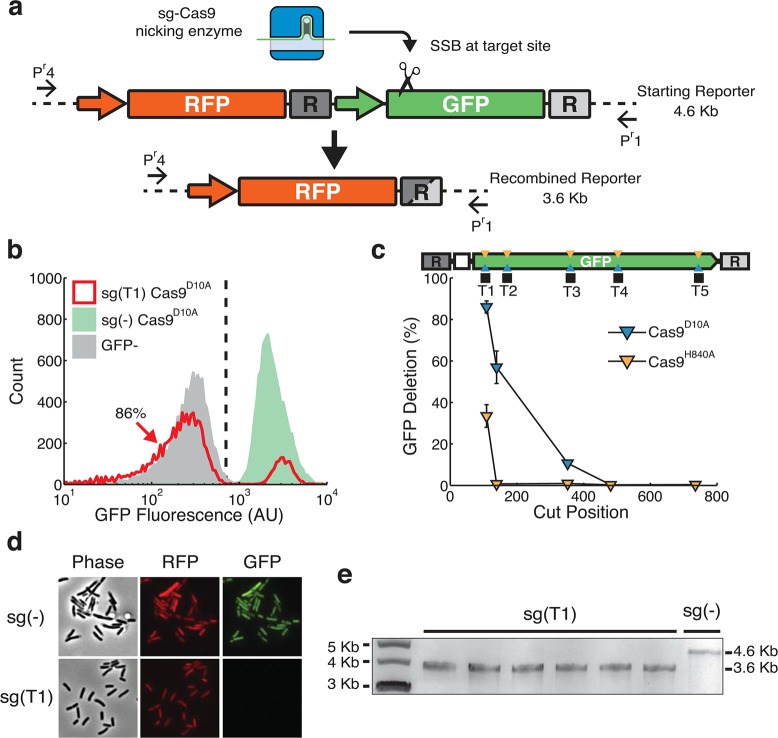
Figure 2.
Nick-induced recombination leads to single gene chromosomal deletion. (a) A diagram of chromosomally integrated dual-fluorescent reporter with internal homologous DNA sequences (R), shown as dark and light gray boxes. sgRNA-guided Cas9 nicking enzyme generates an SSB, illustrated by the scissor. CRISPR induced deletions of GFP results in the joining of two repeats, shown as a gray box with both dark and light shades. Primers (Pr1 and Pr4) flank the dual fluorescence reporter. (b) Flow cytometry screening of pooled sgRNA transformations for homologous recombination. Histograms show population fluorescence distributions for controls sg(−) (green), cells containing RFP with no GFP (gray), and sg(T1) coexpressed with Cas9D10A (red). Vertical dashed line represents threshold for GFP expression. 86% of sg(T1) cells fall to the left of the line, indicating the loss of GFP expression. (c) Percent of flow cytometry estimated GFP deletion for various sgRNAs targeting the noncoding strand using Cas9D10A (blue) and Cas9H840A (orange). Nicking sites within GFP are represented by blue and orange triangles and black squares representing sgRNA target sites. Cut positions are defined as bp from the left repeat (d) Fluorescence microscopy interrogation of dual-fluorescent reporter expression for sg(−) and sg(T1) transformants in E. coli expressing Cas9D10A. GFP expression is undetectable in sg(T1) while RFP expression is similar in both sg(−) and sg(T1). Phase contrast images show cells have normal morphology and RFP expression. (e) Gel-electrophoresis of amplicons using primers Pr1 and Pr4 from transformants for sg(T1) and sg(−). The primers generate a 4.6 kilobase (Kb) amplicon for the initial RFP:GFP site, and following HR the primers generate a 3.6 Kb amplicon. Six out 6 sg(T1) transformants amplify at sizes of approximately 3.6 kilobases, while a sg(−) control template results in banding at 4.6 Kb.