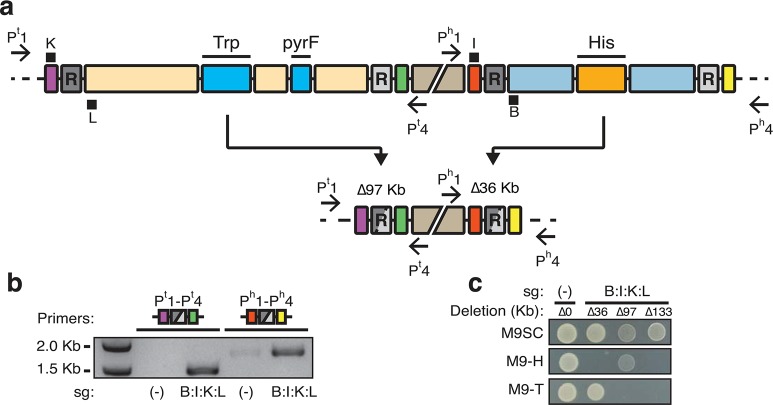
Figure 4.
Multiplex targeted genome remodeling achieves 133 Kb deletion. (a) A schematic view of two separate genomic regions surrounded by direct repeat sequences. Definitions of illustrations are the same as in Figure 3. The middle disjoint beige box represents 670 Kb distance between these two regions. The left region is 97 kilobases and contains the tryptophan biosynthesis operon (TRP) and the pyrF gene (labeled bright blue boxes). Recombination between these repeats results in deletion of 97 Kb (Δ97 Kb) and tryptophan auxotrophy. The remodeled site (purple, gray and green) brings primers Pt1 and Pt4 in into close proximity. sgRNAs K and L target the sequences directly adjacent to the left repeat to this region. In parallel, guides B and I target a separate region containing His (orange box). The position of black squares indicate which strand is targeted by the sgRNA with K and I targeting the + strand and L and B targeting the – strand (see Figure 3 also). Deletion of this region results in a loss of 36 Kb (Δ36 Kb) and histidine auxotrophy. The remodeled sequence (red, gray and yellow) results in close proximity of Ph1 and Ph4. (b) PCR monitoring of HR in Cas9D10A expressing cells. Primers Pt1 and Pt4 detect deletion of 97 Kb (purple, gray and green icon, resulting in a 1.5 Kb amplicon). Ph1 and Ph4 detect recombination across 36 Kb (red, gray and yellow icon, resulting in a 1.8 Kb amplicon). Different sgRNA targets are indicated beneath the gel photo. sg(−) is a guide sequence not matching the bacterial genome. Amplification indicates occurrence of recombination. (c) Resulting phenotypes of isolated E. coli containing different remodeling combinations. Cells isolated containing different deletions replica plated on M9 synthetic complete medium (M9SC), M9 lacking histidine (M9-H), and M9 without tryptophan (M9-T). Vertical columns correspond to different deletions. sg(−) with no genomic deletion (Δ0) serves as the control. sg(B:I:K:L) expressing cells can harbor 36 (Δ36), 97 (Δ97) or combined 133 kilobase (Δ133) deletions. Δ36, Δ97 and Δ133 are auxotrophic histidine, tryptophan and both, respectively.