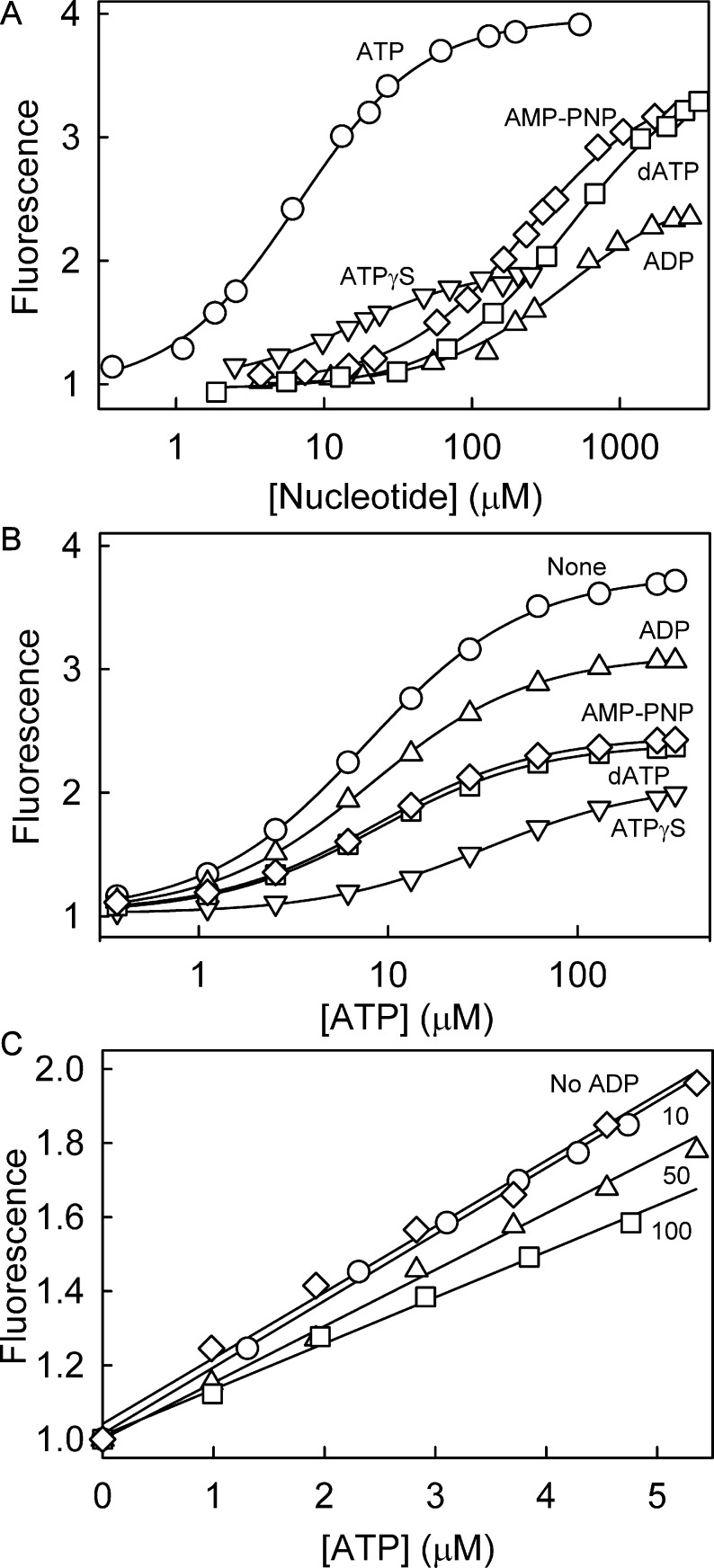
Figure 3.
Nucleotide affinity to Rho-MatB. (A) Titration of ATP (circles), ADP (up triangles), dATP (squares), ATPγS (down triangles), and AMP-PNP (diamonds) to 0.5 μM Rho-MatB in buffer as in Figure 2 at 20 °C. The dissociation constants were obtained using a quadratic binding equation (see Methods) and are listed in Table 1. (B) The effect of different nucleotides on ATP binding. 1 μM Rho-MatB in the absence (circles) and presence of 100 μM ADP, dATP, ATPγS, or AMP-PNP (symbols as above) was titrated with ATP. The apparent dissociation constants for ATP were obtained using the quadratic binding equation. The fitted curves gave the following values for apparent dissociation constant and fluorescence ratio: ADP, 7.7 ± 0.6 μM, 3.0; dATP, 9.1 ± 1.2 μM, 2.3; ATPγS, 38 ± 9 μM, 1.9; AMP-PNP, 8.2 ± 0.4 μM, 2.4. (C) Linearity of response to ATP. 2.5 μM Rho-MatB was titrated with ATP (diamonds). To test the effect of ADP, the fluorescence was measured in the presence of ADP, such that the total nucleotide concentration (ADP + ATP) was constant at 10 μM (circles), 50 μM (triangles), or 100 μM (squares).