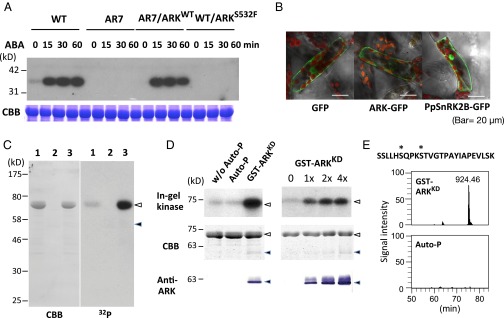
Fig. 3.
ARK as a possible regulator of SnRK2. (A, Upper) In-gel kinase assays of protonemata of WT, AR7, and ARK knock-in lines for the detection of ABA-activated protein kinases. Proteins were electrophoresed on an SDS-polyacrylamide gel containing histone IIIS. After denaturation and renaturation procedures, the proteins were reacted with γ-[32P]ATP. (Lower) Coomassie Brilliant Blue (CBB) staining of ribulose 1,5-bisphosphate carboxylase/oxygenase. (B) The ARK-GFP construct fused to the CaMV35 promoter was introduced into protonema cells of P. patens. Localization of GFP and PpSnRK2-GFP is shown for comparison. (C) Kinase activity of GST-ARKKD on MBP-PpSnRK2B. Recombinant proteins of MBP-PpSnRK2B (1 µg) (lanes 1 and 3) and GST-ARKKD (0.05 µg) (lanes 2 and 3) were reacted with γ-[32P]ATP and electrophoresed on an SDS-polyacrylamide gel, followed by Coomassie Brilliant Blue staining and autoradiography (32P). Closed and open arrowheads indicate the positions of MBP-PpSnRK2B and GST-ARKKD, respectively. (D) In-gel kinase assays of the recombinant MBP-PpSnRK2B (1 µg) with or without autophosphorylation (Auto-P) or phosphorylation by GST-ARKKD (0.05 µg). Positions of molecular-weight markers are shown in kilodaltons. Results of staining with Coomassie Brilliant Blue and immunoblot analysis using the anti-ARK antibody (Anti-ARK) are shown for comparison. Closed and open arrowheads indicate the positions of MBP-PpSnRK2B and GST-ARKKD, respectively. (E) Phosphorylation sites in the activation loop of MBP-PpSnRK2B identified by phosphopeptide mapping. The amino acid sequence and probable phosphorylation sites (asterisks) of the identified phosphopeptide (m/z = 924.46) and its ion-current chromatograms are shown. The MBP-PpSnRK2B protein was either phosphorylated with GST-ARKKD or autophosphorylated (Auto-P) for 15 min. (Details are shown in Fig. S6.)