Figure 4.
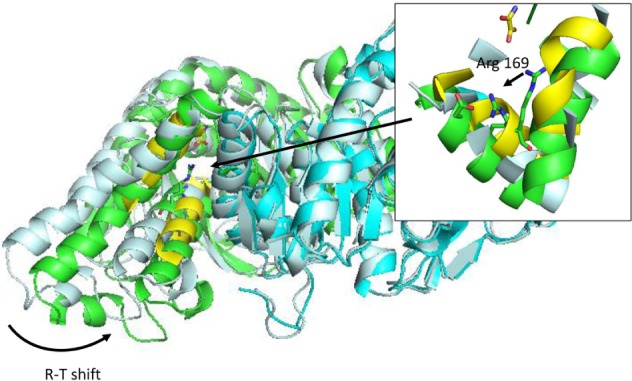
R-T transition on Bacillus stearothermophilus lactate dehydrogenase after 50 ns simulation in explicit solvent.
Notes: Inset: detail of conformational shift of active site substrate-binding Arg 169 side chain. Simulation was initiated from a dimeric model of the protein (1ldn), and allowed to evolve without restrains. Protein system was prepared using MDWeb,91 and simulated using GROMACS,148 at 298 K of temperature, in explicit solvent and periodic boundary conditions (truncated octahedron box). Conformational shift is indicated as R-T shift in the main figure and with an arrow in the inset.
Abbreviation: R-T, relaxed and tense states (as defined by Monod’s model).
