Abstract
The construction of functional biomimetic scaffolds that recapitulate the topographical and biochemical features of bone tissue extracellular matrix is now of topical interest in bone tissue engineering. In this study, a novel surface-functionalized electrospun polycaprolactone (PCL) nanofiber scaffold with highly ordered structure was developed to simulate the critical features of native bone tissue via a single step of catechol chemistry. Specially, under slightly alkaline aqueous solution, polydopamine (pDA) was coated on the surface of aligned PCL nanofibers after electrospinning, followed by covalent immobilization of bone morphogenetic protein-7-derived peptides onto the pDA-coated nanofiber surface. Contact angle measurement, Raman spectroscopy, and X-ray photoelectron spectroscopy confirmed the presence of pDA and peptides on PCL nanofiber surface. Our results demonstrated that surface modification with osteoinductive peptides could improve cytocompatibility of nanofibers in terms of cell adhesion, spreading, and proliferation. Most importantly, Alizarin Red S staining, quantitative real-time polymerase chain reaction, immunostaining, and Western blot revealed that human mesenchymal stem cells cultured on aligned nanofibers with osteoinductive peptides exhibited enhanced osteogenic differentiation potential than cells on randomly oriented nanofibers. Furthermore, the aligned nanofibers with osteoinductive peptides could direct osteogenic differentiation of human mesenchymal stem cells even in the absence of osteoinducting factors, suggesting superior osteogenic efficacy of biomimetic design that combines the advantages of osteoinductive peptide signal and highly ordered nanofibers on cell fate decision. The presented peptide-decorated bone-mimic nanofiber scaffolds hold a promising potential in the context of bone tissue engineering.
Keywords: biomimetic, nanofiber, peptide, bone tissue engineering
Introduction
With the aging population, the occurrence of bone defect associated with tumors, osteoporosis, and osteoarthritis is rising so dramatically that governments around the world have to boost budget to balance the growing expenditure for health care every year.1 Despite the availability of invasive surgeries using metallic/polymeric implants or autografts, it remains a great challenge to completely restore the large bone defects in clinics.2 Stem cell-based tissue engineering (STE), which combines stem cells with a supportive scaffold, offers an enabling alternative for reconstruction and functional recovery of seriously damaged bone tissues.2,3 However, the virtual translation of STE from “bench-to-bedside” still faces many hurdles. One of the major obstacles lies in that the traditional design of scaffold lacks biomimicry and mismatches in tissue morphology, or chemical properties, ultimately leading to the failure of stem cell transplantation.4
Nanofibrous materials fabricated by electrospinning technology have attracted considerable interest in bone tissue regeneration, due to the structural resemblance to the bone tissue extracellular matrix (ECM) in micro- to nanoscale.5 The electrospun nanofibers feature a porous network with a large surface area, rendering an ideal microenvironment for initial cell anchorage and growth in tissue engineering.6,7 Many degradable polymers, with either a synthetic or a natural origin, can be electrospun into nanofibers.5 Nevertheless, the flexibility and shape availability of synthetic nanofibers give them greater potential in the bone regeneration field.5 Through adjusting the electrospinning parameters and collector design, synthetic polymers can be readily processed into various fiber diameters or nanopatterns,3 of which highly ordered nanofibers have been extensively studied for bone tissue engineering toward the treatment of a variety of bone defects8,9 since its topographical feature is similar to the well-aligned organization (eg, long parallel bundles of type I collagen fiber) observed in bone tissue.10 On such aligned substrates, stem cells are arranged in a direction parallel to the nanofibers’ alignment.8 Moreover, increased calcium and collagen production was observed in stem cells cultured on aligned nanofibers as opposed to randomly oriented scaffolds11 because cytoskeletal elongation altered by aligned nanofibers may activate certain osteo-related gene expression via mechanotransduction.11 Given the superior performance in bone formation, highly ordered nanofibers are considered a promising candidate of scaffolds for bone tissue engineering.12
It has been gradually recognized that among various parameters that influence cell–material interactions, not only topographical but also biochemical cues should be considered in the design of scaffold materials that can mimic the chemical and physical features of microenvironments in native tissues.13 Thus, the functionalization of nanofibers with bioactive factors is a critical step to construct bone biomimetic scaffolds, which could simultaneously present multiple cues for the regulation of cell fates. Inspired by the manner of biomolecules tethering to the native ECM, a great deal of efforts have been focused on the surface functionalization of nanofibers with osteogenesis-associated proteins in recent years. Bone morphogenetic proteins (BMPs), members of the transforming growth factor-β superfamily, are widely implicated in the regulation of skeletal development, including proliferation and differentiation of mesenchymal stem cells (MSCs) toward osteogenic fates.3,14 It has been proved that covalent surface immobilization of scaffolds with BMP proteins could convey osteoinductive signals to the local cell microenvironment in a sustained manner.15 However, the complicated multilevel structure of proteins makes them prone to degradation in the physiological environment,16 probably causing the loss of bioactivity. Therefore, short peptide sequences have attracted much attention over the past decade due to their higher structure stability and less immunogenicity compared to large proteins.16 Recently, bone-forming peptide-1 (BFP-1), a novel sequence derived from the immature region of BMP-7, was reported to exhibit more excellent osteogenetic efficacy over BMP-7 protein in vitro and in vivo.14 Accordingly, considerable benefits in osteogenic differentiation of stem cells may derive from a combination of bioinspired nanotopography fabricated with aligned nanofibers and biochemical signaling from BFP-1 peptides.
Guided by these considerations, we developed an innovative bone-mimic BFP-decorated polycaprolactone (PCL, a model synthetic polymer) nanofiber scaffold with highly ordered structure. To surface functionalization of synthetic nanofibers with osteogenic peptides, dopamine chemistry was employed to bridge these two stimulators in the scaffold design. To the best of our knowledge, this is the first report that the presented biomimetic materials, which combine aligned nanofibers and osteogenic peptides, are designed to promote osteogenesis for bone tissue engineering. To evaluate the cytocompatibility and osteogenic activity of these materials, the adhesion, growth, and differentiation of human mesenchymal stem cells (hMSCs) cultured on the peptide-decorated nanofibers were deeply investigated in vitro.
Materials and methods
Materials
PCL (molecular weight [MW] =80,000 g/mol) and dopamine hydrochloride were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Hexafluoro-2-propanol and Tris–HCl were purchased from Aladdin Reagent Co, Ltd (Shanghai, People’s Republic of China). α-Minimum Essential Medium (α-MEM medium), fetal bovine serum, phosphate-buffered saline (PBS), trypsin–ethylenediaminetetraacetic acid, and penicillin–streptomycin were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Distilled water (DW) was obtained from the Milli-Q Plus System (EMD Millipore, Billerica, MA, USA). The peptide (KGGQGFSYPYKAVF-STQ sequence) was synthesized by China Peptides Co, Ltd (Shanghai, People’s Republic of China). All other chemicals and solvents were purchased from Sigma-Aldrich Co.
Preparation of PCL nanofibers and surface functionalization
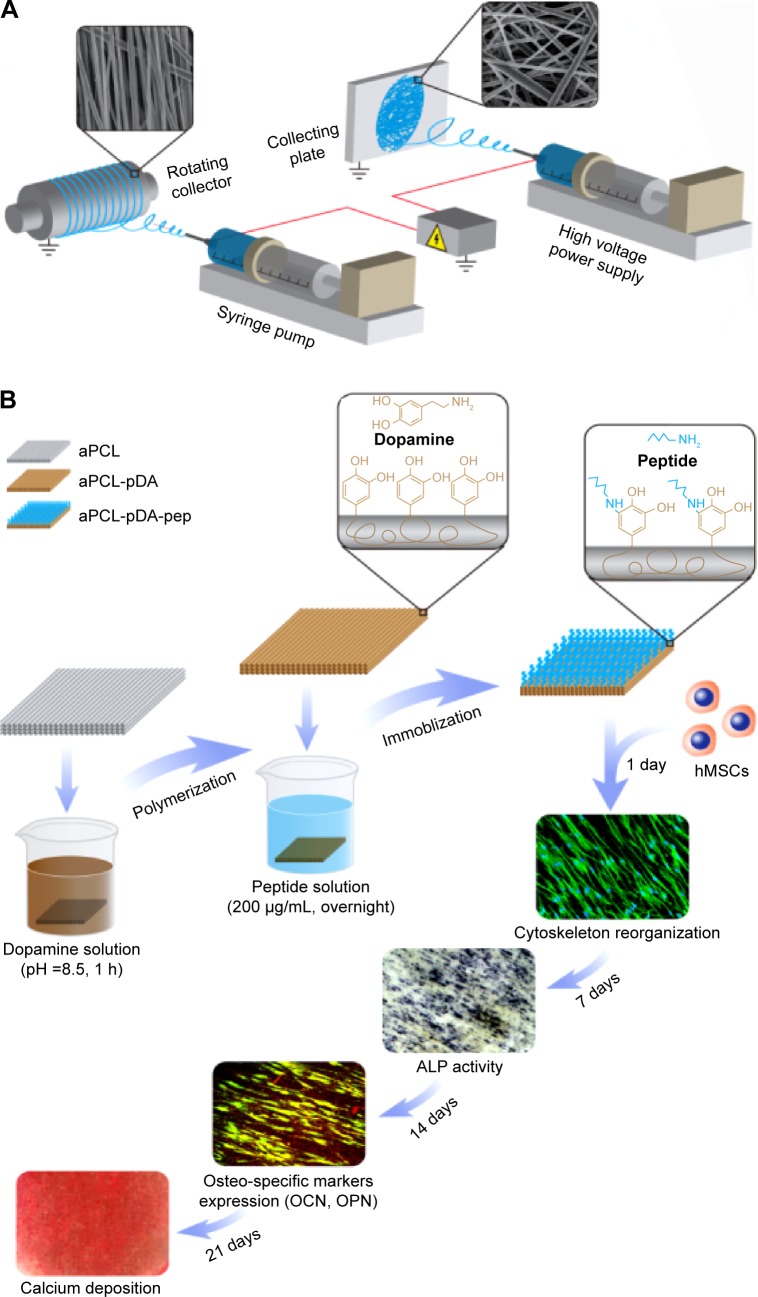
PCL nanofibers were fabricated using a commercial electro-spinning equipment (SS-2535; YongKang Technology Co, Ltd, Beijing, People’s Republic of China) (Figure 1A). In brief, PCL solution (8 wt%) prepared in hexafluoro-2-propanol was fed into a plastic syringe with a stainless-steel blunt needle (23 G). Randomly oriented and aligned nanofibers were collected on a flat collector (applied voltage, 14 kv; feed rate, 1 mL/h; collection distance, 10 cm) and a high-speed rotating collector (applied voltage, 10 kV; solution feed rate, 2 mL/h; collection distance, 5 cm), respectively. The as-prepared fibers were dried in vacuum for 1 day to remove residual organic solvent. For surface functionalization, the nanofibers were pre-immersed in dopamine solution (2 mg/mL, 10 mM Tris buffer, pH 8.5) and gently shaken for 1 hour at room temperature (RT), as previously described.17 The polydopamine (pDA)-coated substrates were then thoroughly rinsed with DW and further reacted with BFP-1 peptide (KGGQGFSYPYKAVFSTQ sequence) solution in PBS overnight at 37°C (Figure 1B, and Figures S1 and S2). The codes for the prepared samples are presented in Table 1.
Figure 1.
Schematic illustration of (A) fabrication of electrospun nanofibers and (B) surface functionalization of PCL nanofibers with BFP-1 peptides for directing hMSCs osteogenic commitment.
Abbreviations: aPCL, aligned PCL nanofibers; PCL, polycaprolactone; BFP-1, bone-forming peptide-1; hMSCs, human mesenchymal stem cells; pDA, polydopamine; pep, peptide; ALP, alkaline phosphatase; OCN, osteocalcin; OPN, osteopontin; h, hour.
Table 1.
PCL nanofiber formulations
| Sample code | Nanofiber orientation | pDA coating (h) | Peptide solution (μg/mL) |
|---|---|---|---|
| R-pDA | Random | 1 | – |
| R-pDA-pep | Random | 1 | 200 |
| A-pDA | Aligned | 1 | – |
| A-pDA-pep | Aligned | 1 | 200 |
Abbreviations: h, hour; PCL, polycaprolactone; pDA, polydopamine; pep, peptide; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber.
The surface topography of nanofibers was observed by a scanning electron microscope (SEM, S-4800; Hitachi Ltd, Tokyo, Japan) and a white-light interferometer (WLI, Contour GT-X8; Bruker Corporation, Billerica, MA, USA). Furthermore, the surface chemistry of nanofibers was characterized by water contact angle measurement (SL200B; KINO Industry, New York, NY, USA), Raman spectroscopy (Renishaw, Gloucestershire, UK), and X-ray photoelectron spectroscopy (XPS, AXIS Ultra; Kratos Analytical, Ltd, Manchester, UK).
Cell culture and osteogenic differentiation
All protocols involving human cells were approved by the Institutional Review Board of Peking University (Beijing, People’s Republic of China). hMSCs, purchased from ScienCell Research Laboratories (Carlsbad, CA, USA), were subcultured in α-MEM containing fetal bovine serum (10%) and penicillin–streptomycin (1%) under standard culture condition. The growth media were changed every 2 days. To evaluate cell responses to materials, the nanofiber matrices were punched into round shape and sterilized with ethanol (75%) for 1 hour before cell culture experiment. hMSCs were plated onto fibers at a density of 2.0×104 cells/cm2. After 1 day, the media were replaced with osteoinductive media (OM), which was prepared by adding ascorbic acid (50 μg/mL), β-glycerophosphate (10 mM), and dexamethasone (100 nM) to the growth medium.
Imaging cell morphology using SEM
At predetermined time points, the samples were washed twice with PBS (1×), fixed in glutaraldehyde (2.5%) for 2 hours, and then dehydrated with gradient ethanol (30%–100%). Each sample was dried with hexamethyldisilazane for 15 minutes prior to sputter coating with gold for SEM observation.
CCK-8 assay
Cell adhesion and proliferation on functionalized nanofibers were evaluated by Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. At preset time points, the culture medium was removed and the samples were washed with PBS. Afterward, CCK-8 reagent (10%) in culture media was added to each well. After 2-hour incubation, supernatant (100 μL) from each well was transferred to a 96-well plate for the measurement of absorbance value at 450 nm using a microplate reader.
Live/dead assay
At 1, 3, and 5 days, after washing twice with PBS, cells on the samples were incubated with serum-free culture media containing fluorescein diacetate (FDA, 5 μg/mL) and propidium iodide (PI, 20 μg/mL) at RT for 5 minutes in dark. Images were captured using a laser scanning confocal microscope to visualize FDA-stained live cells and PI-stained dead cells.
Alkaline phosphatase assay
Alkaline phosphatase (ALP) activity of hMSCs was evaluated by an assay reagent kit (Nanjing Jiancheng Bioengineering Institute, Jiangsu, People’s Republic of China). On days 7 and 14, the supernatant was removed and Triton X-100 solution (1%, 100 μL) was added into each well for cell lysis. After 1-hour incubation at 4°C, 30 μL of cell lysate from each well was transferred to a 96-well plate, cultivated with carbonated buffer solution (50 μL, pH 10) and 4-amino-antipyrine solution (50 μL) at 37°C for 15 minutes. Then potassium ferricyanide (150 μL) was added into the above solution, and the production of p-nitrophenol was determined by the absorbance at 405 nm. For normalization, the total protein content was measured by a bicinchoninic acid protein assay kit (Thermo Fisher Scientific), and ALP activity was expressed as U/mg protein. Furthermore, enzyme histochemistry staining was performed to visualize ALP expression using a 5-bromo-4-chloro-3-indolylphosphate/nitro blue tetrazolium ALP color development kit (Beyotime Biotechnology, Shanghai, People’s Republic of China) according to the manufacturer’s instructions.
Alizarin Red S staining
After 21 days of culture, cells were fixed in ethanol (95%) for 30 minutes, followed by washing thrice with DW. Then, the fixed cells were stained with Alizarin Red S (ARS) (2%, pH 4.2) for 10 minutes at RT and thoroughly washed with DW for imaging using scanner. To quantify the orange-red coloration of ARS, the colorant on the samples was eluted with cetylpyridinium chloride (10%) solution for 1 hour at RT, and the eluate (200 μL) was transferred to a 96-well plate for the measurement of absorbance value at 550 nm using a microplate reader.
RNA isolation and real-time polymerase chain reaction
The expression of osteogenic-specific genes encoding Runt-related transcription factor 2 (Runx2), type I collagen alpha 1 (Col1a1), and osteocalcin (OCN) at messenger RNA level was characterized on day 14 by real-time polymerase chain reaction (RT-PCR) (Thermo Fisher Scientific). Total RNA was isolated with TRIzol and reverse transcribed with RevertAid™ First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania). The resultant complementary DNA was applied for the PCR using SYBR Green. The relative level of each target gene was normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) using a comparative cycle threshold (ddCt) method. Primers used in this study were listed in Table S1.
Immunocytochemistry
At preset time points, cells on the samples were fixed with 4% paraformaldehyde for 10 minutes and permeabilized with Triton X-100 (0.1%) for 5 minutes. After blocking with bovine serum albumin solution (1%) for 2 hours, the samples were incubated overnight at 4°C with primary antibodies: mouse monoclonal anti-human OCN (1:100 dilution; Abcam, Cambridge, MA, USA), rabbit polyclonal anti-human osteopontin (OPN, 1:500 dilution; Abcam), and mouse monoclonal anti-human paxillin (1:1,000 dilution; BD Biosciences, San Jose, CA, USA). Afterward, cells were washed thrice with PBS and incubated for 1 hour at RT with the appropriate secondary antibody: fluorescein isothiocyanate-488 goat anti-rabbit (1:1,000; Cell Signaling Technology, Boston, MA, USA) or tetramethylrhodamine isothiocyanate-543 goat anti-mouse (1:1,000; Cell Signaling Technology). Subsequently, 4′,6-diamidino-2-phenylindole (10 μg/mL) and fluorescein isothiocyanate–phalloidin (5 μg/mL) were used for counterstaining of cell nuclei and F-actin, respectively. Fluorescence signals were captured under confocal microscope (Carl Zeiss Meditec AG, Jena, Germany).
Western blotting
hMSCs were harvested on day 14 and lysed by radio immunoprecipitation assay buffer containing protease inhibitor cocktail. Protein concentration was determined by a bicinchoninic acid protein assay kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Protein samples (25 μg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10%) and transferred to poly vinylidene fluoride membranes (EMD Millipore). After blocking with skimmed milk (5%) in TBS containing 0.05% Tween-20 (TBST) for 1 hour, the membranes were incubated overnight at 4°C with primary antibodies: anti-β-actin (1:1,000; Cell Signaling Technology), anti-GAPDH (1:1,000; Cell Signaling Technology), anti-human Runx2 (1:3,200; Cell Signaling Technology), anti-human Col1a1 (1:1,000; Santa Cruz Biotechnology Inc., Dallas, TX, USA), and anti-human paxillin (1:1,000; BD Biosciences), followed by washing thrice with TBST. Afterward, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:10,000; Cell Signaling Technology) for 1 hour at RT. After washing thrice with TBST, the horseradish peroxidase reaction product was detected by ECL Western Blotting kit.
Statistical analysis
All experiments were performed in triplicate, and the results were expressed as mean ± standard deviations. One-way analysis of variance and Tukey’s post hoc test were applied to statistically evaluate significant differences among groups by means of SPSS software (version 13.0; SPSS Inc., Chicago, IL, USA). A value of P<0.05 was considered statistically significant.
Results and discussion
Microstructural characteristics of functionalized nanofibers
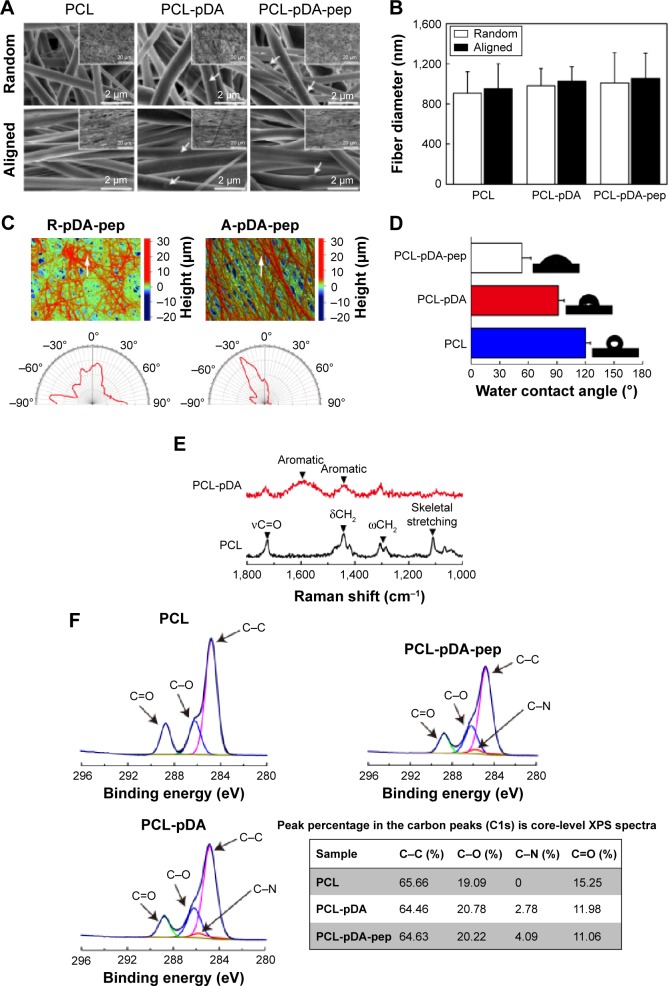
As shown in SEM images (Figure 2A), the pure aligned and randomly oriented nanofibers exhibited smooth and uniform morphology without bead defects. However, some nanosized particulates and incremental discrepancies in surface roughness were observed on nanofibers after surface modification of pDA and peptides, even though no significant differences in fiber diameters were detected among groups (P>0.05; Figure 2B). These findings were quantitatively confirmed by WLI analysis as shown in Table S2 (P<0.05), suggesting pDA and peptides could roughen the surface of the substrates.18 Moreover, the orientation degree of nanofibers was analyzed by WLI (Figure 2C). Most nanofibers in the randomly oriented substrates showed a broad distribution of ±90° with respect to the reference axis, while the aligned ones presented an intensive distribution with the majority of nanofibers within −30°, implying potential differences in the cell fate commitment associated with nanofiber orientation.
Figure 2.
Surface characterization of functionalized nanofibers.
Notes: (A) SEM images of peptide-decorated nanofibers. The white arrows represent the pDA aggregate particulates. The corresponding SEM images obtained at low magnification are shown as insets. (B) Fiber diameter measurement of nanofibers. (C) Representative surface topographical images of peptide-decorated nanofibers. The white arrows indicate the reference direction for orientation evaluation. The compass images represent the orientation distribution of peptide-decorated nanofibers with random and aligned orientation. (D) Water contact angle and representative images of water droplet on PCL nanofibers with different surface chemistries. (E) Raman spectra analysis of PCL and pDA-coated nanofibers. (F) High-resolution carbon peaks (C1s) from PCL nanofibers with different surface chemistries. The table shows the peak percentage in the carbon peaks (C1s) core-level XPS in different nanofibers.
Abbreviations: SEM, scanning electron microscope; pDA, polydopamine; PCL, polycaprolactone; XPS, X-ray photoelectron spectroscopy; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide.
Construction of biomimetic materials that simulate the microstructural features of bone ECM is critical to the advancement of scaffold design targeted at an efficient control of stem cell fate. Recently, diverse bone-mimic nanopatterned platforms for directed osteogenic differentiation were reported.2,19 For instance, work done by Kim et al19 demonstrated that hMSCs grown on the polyurethane acrylate substrates with bone-like nanoscale groove-shaped topography exhibited significant upregulation of osteo-specific markers expression compared to those on unpatterned surfaces. Despite a straightforward approach for in vitro research, such reported patterned substratum, merely a simplified platform, cannot create three-dimensional architecture with desired topographies such as fibrous building blocks and highly interconnected porous structures.3,20 Furthermore, the fabrication of these substrates requires complex processing procedures such as photolithography and electrochemical etching, limiting their applicable potential in clinics. On the contrary, the nanofibrous matrices fabricated by electrospinning process have gained increasing interest over the past decade, mainly due to the structural similarity to the tissue ECM, the processing availability to a wide range of materials, as well as simple setup and operation at low cost.5 These intriguing features endow electrospun nanofibers with greater promise as scaffolds for bone tissue engineering.
Surface characterization of functionalized nanofibers
Water contact angle is a convenient approach to evaluate the hydrophilic–hydrophobic attributes of nanofiber surfaces. It is documented that pDA coating could improve the hydrophilicity properties of polymeric biomaterials.21 Consistent with previous reports, the water contact angle on the aligned PCL nanofibers coated with pDA for 1 hour decreased from 120.1°±5.3° to 91.3°±6.5° (Figure 2D), due to the hydrophilicity of dopamine molecules with a catecholamine group. Moreover, the immobilization of peptides on the pDA-coated nanofiber surface further decreased the water contact angle to 53.1°±9.6°, which implies the successful conjugation of peptides onto nanofibers (P<0.05).
To further validate the results of water contact angle, Raman spectra and XPS were obtained as shown in Figure 2E and F, respectively. Typical bands in the range of 1,725 cm−1 (νC=O), 1,419–1,470 cm−1 (δCH2), 1,282–1,308 cm−1 (ωCH2), and 1,112 cm−1 (skeletal stretching) for PCL nanofibers were detected in Raman spectra,22 whereas pDA-coated sample displayed additional peaks at ~1,590 cm−1 and 1,440 cm−1, which correspond to the deformation and stretching of catechol, respectively,21 indicating the successful deposition of pDA onto nanofibers. In XPS (Figure 2F), peaks at 284.8 eV (C–C), 286.2 eV (C–O), and 288.8 eV (C=O) were observed on PCL nanofibers, which could be ascribed to the presence of esters in the main backbone of the polymer,23 whereas a signal for C–N bond (286 eV) was recorded on both pDA-coated and peptide-decorated nanofibers. Quantification of the intensity of C–N bond revealed a gradual enhancement on the pDA-coated and peptide-decorated PCL nanofiber surfaces (Figure 2F), proving the presence of pDA and peptides on the functionalized nanofibers.
Surface functionalization is a straightforward and robust approach to regulate cell behaviors. To modify the interfacial attributes of synthetic nanofibers, physical adsorption and chemical covalent conjugation have been widely explored in recent years.6 Physical adsorption is the simplest and drug-friendly strategy for loading biomolecules on the nanofibrous mesh. However, the binding affinity is highly dependent on the type of materials, and long-term stability is not guaranteed.24 Thereby, chemical covalent conjugation is favored over physical adsorption in tissue engineering because the covalently grafted molecules are not easily leached out from the surface-modified nanofibers for long-term application.6 On the other hand, conventional chemical conjugation approaches often require complicated chemical steps, which may lead to denaturation or partial inactivation of the grafted molecules in the harsh chemical environment, limiting their applicability in clinic.24 Recently, a facile, green, and potent surface modification strategy for immobilizing bioactive factors via simple dip-coating with dopamine solution was reported.25 Dopamine, a mussel-inspired monomer, could initiate oxidative self-polymerization and form a thin adherent polymer film on material surfaces in weak alkaline solution, which enable easy conjugation of biomolecules containing thiols or primary amines moieties via Schiff base reactions or Michael addition,25 thereby holding a promising potential in the surface engineering of bone-mimic nanofiber materials.
Cell adhesion and spreading on functionalized nanofibers
Adhesion and spreading are the first event of cell response to a substrate, which plays a profound role in determining stem cell fate such as self-renewal and differentiation.26 Therefore, early adhesion and spreading of hMSCs on functionalized nanofibers were evaluated under serum-free conditions in order to eliminate the disturbance of soluble ECM adhesion proteins from serum (Figure S3).
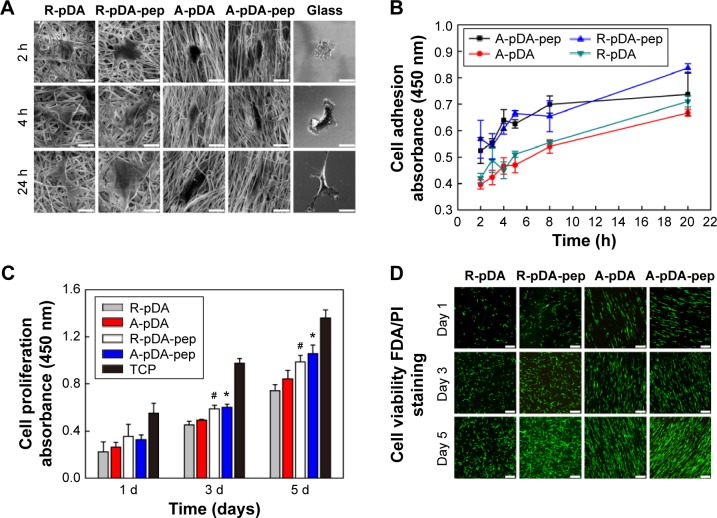
As shown in SEM (Figure 3A), time-dependent shape of hMSCs was observed on functionalized nanofibers. At 2 hours after seeding, cells on control glass exhibited fried egg-like morphology without pseudopodium, but on nanofiber surfaces, regardless of fiber orientation, short lamellipodia were observed at the periphery of cells, showing favorable affinity of nanofiber substrates to cell adhesion. In addition, compared to hMSCs in R-pDA and A-pDA groups, cells in R-pDA-pep and A-pDA-pep groups displayed improved spreading morphology, highlighting the significance of BFP-1 peptides for early cell spreading on nanofibers. In consistent with SEM observation, CCK-8 data (Figure 3B) showed that at 2 hours, the number of cells in R-pDA-pep and A-pDA-pep groups was more than that in R-pDA and A-pDA groups (P<0.05), respectively. However, no significant differences were showed between R-pDA and A-pDA groups, or between R-pDA-pep and A-pDA-pep groups (P>0.05), which was further confirmed by laser scanning confocal microscope photographs (Figure S4), suggesting the little effect of fiber orientation on early cell attachment. Similar trend was found at 4 hours after seeding. At these time points, hMSCs on randomly oriented nanofibers presented noticeable polygonal profile without a preferential direction, whereas cells on aligned fibers showed spindle-like shape and arrayed along the fiber axis, which recapitulated the aboriginal cell shape and orientation in bone tissue.27 In contrast, hMSCs on glass only began to extend lamellipodia. At 24 hours after seeding, hMSCs had been sufficiently spread out in all groups. Our results showed that surface modification of BFP-1 peptides may contribute to the adhesion and spreading of hMSCs on pDA-coated nanofibers.
Figure 3.
hMSCs’ adhesion, proliferation, and viability on surface-functionalized PCL nanofibers.
Notes: (A) SEM observation and (B) CCK-8 assay of adhering hMSCs on samples in serum-free growth media. Scale bars indicate 15 μm. (C) The proliferation of hMSCs on functionalized nanofibers for 1, 3, and 5 days. #Compared with R-pDA, P<0.05; *compared with A-pDA, P<0.05. (D) Viability of hMSCs on functionalized nanofibers labeled by FDA/PI staining. Live cells: green; dead cells: red. Scale bars indicate 100 μm.
Abbreviations: SEM, scanning electron microscope; CCK-8, Cell Counting Kit-8; hMSCs, human mesenchymal stem cells; pDA, polydopamine; FDA, fluorescein diacetate; PI, propidium iodide; h, hours; TCP, tissue culture plate; PCL, polycaprolactone; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide.
Cell proliferation and viability on functionalized nanofibers
Successful engineered scaffolds are not only supportive of cell adhesion but also improve cell proliferation. Clearly, Figure 3C shows that all test groups exhibited good time-dependent cell growth. However, lower optical density value was observed in all experimental groups compared with tissue culture plate (TCP) control during incubation (P<0.05), indicating somewhat negative influence on cell growth of nanofibers, regardless of orientation and surface chemistries. The possible explanation is that the scaffold in pursuit of osteogenic differentiation may inhibit cell proliferation to some extent.28 Still, A-pDA-pep and R-pDA-pep groups displayed higher optical density value than A-pDA and R-pDA groups (P<0.05), respectively, after incubating for 3 and 5 days, even though no statistical difference was showed between A-pDA-pep and R-pDA-pep samples as well as between A-pDA and R-pDA samples (P>0.05) during culture period. In agreement with the results of lactate dehydrogenase (Figure S5) and proliferation assay, FDA/PI staining (Figure 3D) displayed that hMSCs grew well in each group during 5-day cultivation, and much more viable cells were captured in R-pDA-pep and A-pDA-pep groups on days 3 and 5, compared to R-pDA and A-pDA groups, respectively, suggesting that nanofiber orientation has little impact on cell growth, but proper surface modification with BFP-1 peptides could endow the nanofibers with more desirable cytocompatibility.
hMSCs osteogenic differentiation under osteoinductive conditions
To evaluate the osteogenic bioactivity of peptide-decorated nanofibers with aligned orientation, the extent of osteogenesis in hMSCs on functionalized nanofibers with different orientation and surface chemistries was assessed under osteoinductive condition.
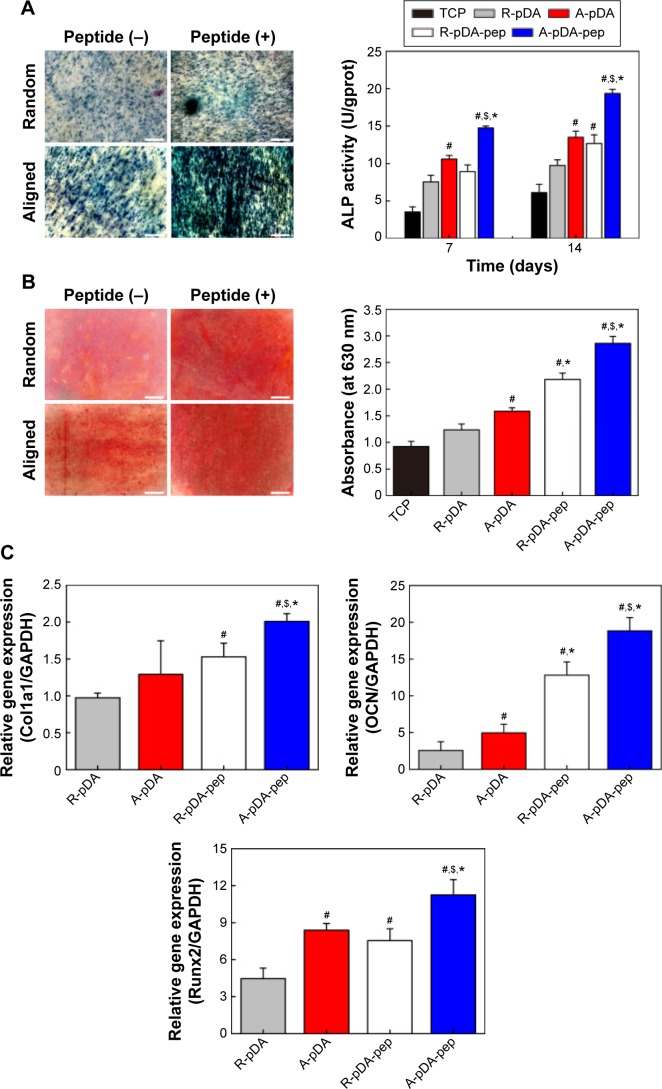
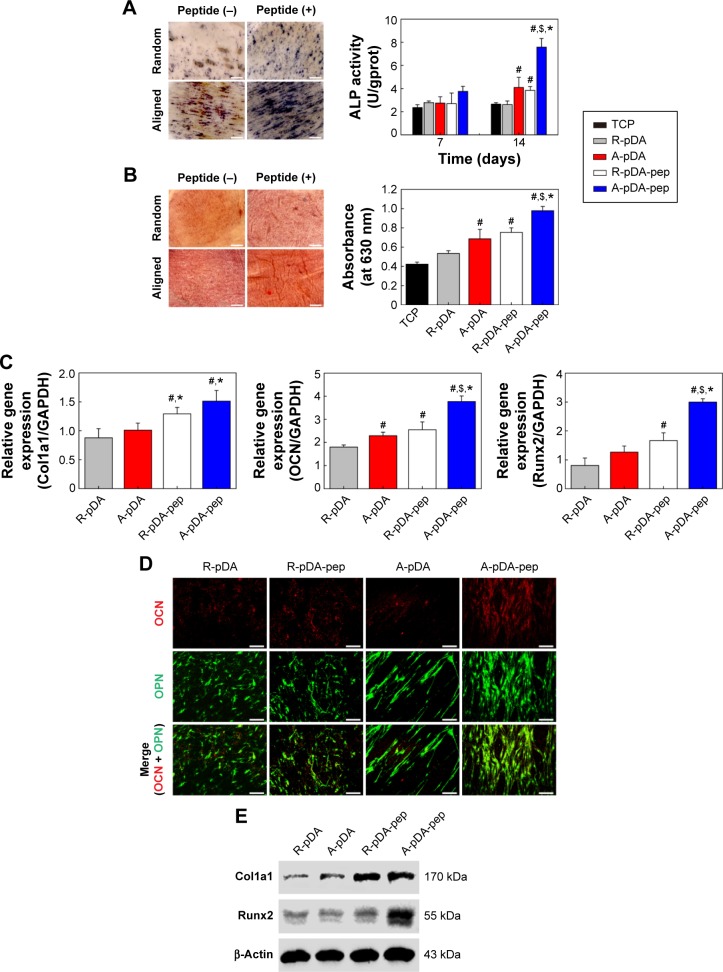
Among the major osteogenic hallmarks, the upregulation of ALP activity is considered a key event occurring during the early time points of osteogenesis.1 As shown in Figure 4A, compared to control TCP group, all test groups demonstrated an enhancement in ALP activity. A-pDA group exhibited higher ALP activity in both time points than R-pDA group (P<0.05), indicating the positive influence of aligned nanofibers on the osteogenic differentiation of stem cells. Furthermore, an increase in ALP activity was detected in A-pDA-pep and R-pDA-pep samples with a peak at 14 days due to the immobilization of BFP-1 peptides on the nanofiber surfaces. This finding suggests that the presence of peptides on the nanofiber surfaces is capable of triggering an upregulation of ALP, correlated with the first checkpoint for osteogenic differentiation. It is noteworthy that more than a 1.5-fold increase in ALP activity was observed in A-pDA-pep samples compared to R-pDA-pep samples on day 14, highlighting a significant synergistic effect between aligned nanofibers and peptides on the ALP expression of hMSCs, which was further qualitatively confirmed via enzyme histochemistry staining of ALP at 14 days (Figures 4A and S6).
Figure 4.
The effect of functionalized nanofibers on osteogenic differentiation of hMSCs under osteoinductive condition.
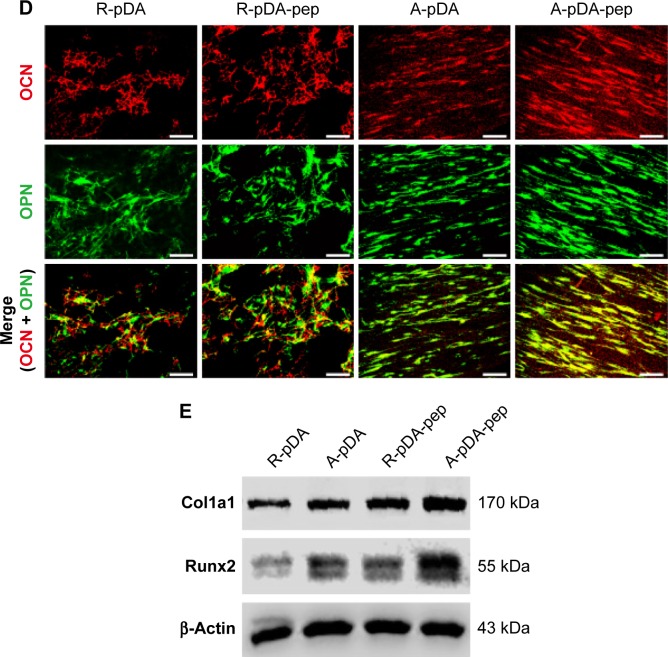
Notes: (A) Representative staining of ALP on day 14 and determination of ALP activity at 7 and 14 days. (B) ARS staining and determination of calcium deposition on day 21. (C) RT-qPCR analysis for osteo-specific genes on day 14. (D) Representative immunofluorescent images of OCN (green) and OPN (red) in different groups on day 14. Scale bars indicate 100 μm. (E) Western blot analysis of Col1a1 and Runx2 expression in hMSCs on different samples. #Compared with R-pDA, P<0.05; *compared with A-pDA, P<0.05; $compared with R-pDA-pep, P<0.05.
Abbreviations: hMSCs, human mesenchymal stem cells; ALP, alkaline phosphatase; ARS, Alizarin Red S; RT-qPCR, real-time quantitative polymerase chain reaction; OCN, osteocalcin; OPN, osteopontin; Col1a1, type I collagen alpha 1; Runx2, Runt-related transcription factor 2; pDA, polydopamine; TCP, tissue culture plate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PCL, polycaprolactone; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide.
The production of calcium deposit at late stage of differentiation is another critical indicator for osteogenic efficiency of hMSCs.29 As the osteogenic culture of hMSCs on functionalized nanofibers progressed to 21 days, cells aggregated together and formed bone-like structures that were stained for ARS (Figure 4B). Similar to the trend observed with ALP activity, more calcium nodules were detected on all nanofibers in comparison with TCP surface, suggesting the osteogenic advantages of nanofibers over TCP surface. Furthermore, it is clearly seen that the aligned nanofibers induced denser red staining than random nanofibers. Due to the conjugation of peptides on nanofibers, a significant increment in the amount of mineralized matrix was observed in both R-pDA-pep and A-pDA-pep groups. Particularly, hMSCs on peptide-conjugated aligned nanofibers showed the highest production of calcium nodules among all groups (P<0.05). In addition, it is noteworthy that R-pDA-pep group induced more production of calcium deposition than A-pDA group by day 21 (P<0.05). This result suggests that biochemical cues arising from BFP-1 peptides may play a greater role in the osteogenic differentiation of hMSCs than topographical cues imparted by aligned nanofibers. However, an opposite result was observed in the ALP assay. The hMSCs on A-pDA samples presented relatively higher ALP activity compared to those on R-pDA-pep samples at 7 days according to the quantitative measurement of ALP (P<0.05). The reasonable explanation is that the impact of topography cues on osteoblast differentiation may commence in the earlier stage compared to biochemical cues, which attributes to the dramatic cell response to aligned fibers since the moment the cells are in contact with substrates, resulting in the cytoskeletal reorganization in the short term and subsequent activation of intracellular signaling associated with osteogenesis. But as far as final osteogenic efficiency is concerned, BFP-1 peptides may exert stronger influence on the cell fate in comparison with aligned fibers.
To better understand the cellular interactions with functionalized nanofibers at transcript level, the expressions of osteo-specific genes were analyzed on day 14 (Figure 4C). The fold change of target genes was normalized to undifferentiated control hMSCs. Of all the osteo-related genes, OCN is regarded as the most specific marker for the mature osteoblast and mineralization during the course of osteogenesis.30 It accumulates in the calcified bone due to its high affinity to hydroxyapatite crystals. As shown in Figure 4C, OCN gene was upregulated in A-pDA group in comparison with R-pDA group (P<0.05). When cells were cultured with peptide-decorated nanofibers, even higher expression of OCN was detected compared to peptide-unmodified group, irrespective of fiber orientation (P<0.05). It is noted that hMSCs in A-pDA-pep group showed a nearly 20-fold upregulation in expression of OCN, which was more than nearly 1.5 times of the expression observed in R-pDA-pep group. Clearly, the modification of BFP-1 peptides on nanofibers could further elevate the OCN expression, especially when applied to the aligned fibers, which is also confirmed along with another osteo-specific protein OPN in immunofluorescence test (Figure 4D). Moreover, the expression profiles of gene Runx2, which is associated with the initiation of osteoblast differentiation,31 and Col1a1, which is involved in the biosynthesis of ECM, correlated well with the change in OCN, and Western blot analysis appeared to support this result at protein level (Figure 4E).
hMSCs osteogenic differentiation under non-osteoinductive conditions
hMSCs could experience spontaneous osteogenic differentiation in the OM due to the presence of osteoinducing factors, dexamethasone, β-glycerophosphate, and ascorbic acid. To circumvent the interference of OM in the intrinsic osteoinductive activity of peptide-functionalized nanofibers, we concomitantly assessed the differentiation of hMSC in normal media via the same set of experiments performed under osteoinductive condition.
As shown in Figures 5A, B and S6, hMSCs on TCP surface maintained an undifferentiated state under non-osteoinductive condition because of extreme low level of ALP activity in cells during early time points and little production of calcium nodules after 21 days of culture. However, when hMSCs were seeded on peptide-decorated aligned nanofibers, the ALP expression in cells was significantly elevated at 14 days and exhibited highest level among groups even though the activity decreased under non-osteoinductive condition (Figure 5A). Similar to the trend of ALP expression, hMSCs in A-pDA-pep group formed much more bone-like nodules compared to other three groups on day 21 (Figure 5B). RT-PCR results showed that when BFP-1 peptides were grafted to aligned nanofiber surfaces, a significant upregulation of OCN and Runx2 genes was observed in hMSCs after 14 days of coculture (Figure 5C). The results of immunofluorescent staining and Western blot corroborated the similar effect as well. Due to the surface modification of BFP-1 peptides to aligned nanofibers, a manifest enhancement in the deposition of OCN and OPN was viewed in confocal images (Figure 5D). Western blotting analysis also revealed that the expression of Runx2 protein in A-pDA-pep group was much stronger than that in R-pDA-pep group even though the production of Col1a1 protein between them was abutting, corresponding to the data from RT-PCR (Figure 5E). Taken together, the osteogenic capacity of hMSCs cultured in normal media is not comparable to those in OM. However, the combined application of aligned nanofibers and BFP-1 peptides showed predominant activation in the osteogenic differentiation of hMSCs even without addition of soluble osteoinducing factors.
Figure 5.
The effect of functionalized nanofibers on osteogenic differentiation of hMSCs under non-osteoinductive condition.
Notes: (A) Representative staining of ALP on day 14 and determination of ALP activity at 7 and 14 days. (B) ARS staining and determination of calcium deposition on day 21. (C) RT-qPCR analysis for osteo-specific genes on day 14. (D) Representative immunofluorescent images of OCN (green) and OPN (red) in different groups on day 14. Scale bars indicate 100 μm. (E) Western blot analysis of Col1a1 and Runx2 expression in hMSCs on different samples. #Compared with R-pDA, P<0.05; *compared with A-pDA, P<0.05; $compared with R-pDA-pep, P<0.05.
Abbreviations: hMSCs, human mesenchymal stem cells; ALP, alkaline phosphatase; ARS, Alizarin Red S; RT-qPCR, real-time quantitative polymerase chain reaction; OCN, osteocalcin; OPN, osteopontin; Col1a1, type I collagen alpha 1; Runx2, Runt-related transcription factor 2; pDA, polydopamine; TCP, tissue culture plate; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PCL, polycaprolactone; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide.
Enhanced focal adhesion of hMSCs by aligned nanofibers
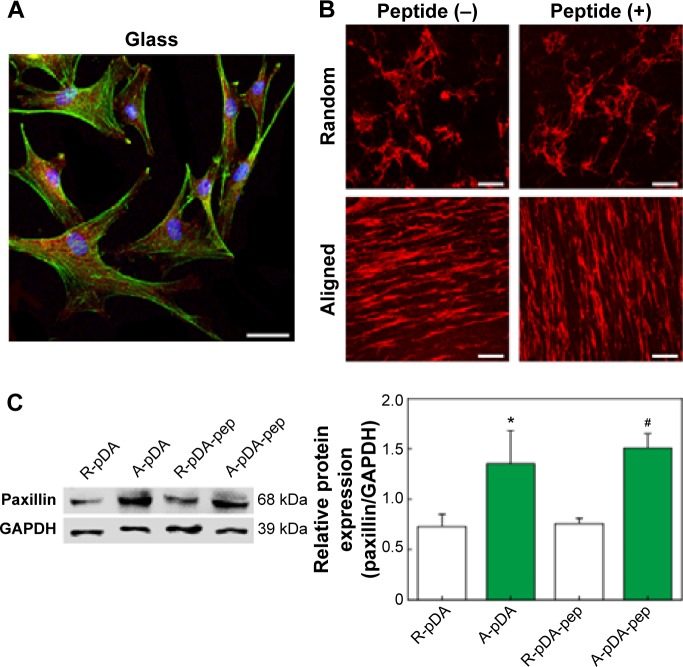
The enhanced osteogenic differentiation of hMSCs on functionalized nanofibers with highly ordered structures may be correlated to the upregulation of focal adhesion proteins by the stimulation of nano- or microscale topographical features emanating from cell culture substrates.26 Thereby, the expression of paxillin, a typical focal adhesion adaptor protein found at the interface between the plasma membrane and the actin cytoskeleton (Figure 6A),32 was investigated on day 7 by immunofluorescent staining and Western blot analysis. Cells were cultured under serum-containing condition. As shown in Figure 6B, the paxillin in hMSCs was polarized along the direction of aligned nanofibers compared to the negligible polarization on the randomly oriented nanofibers, which accompanied with cytoskeletal alteration influenced by fiber orientation. In addition, the distribution area of fluorescence signal emitted from paxillin on A-pDA and A-pDA-pep nanofibers was larger than that on R-pDA and R-pDA-pep nanofibers, respectively, indicating the powerful influence of aligned nanofibers on the focal adhesion formation of hMSCs. Western blot analysis clearly revealed that nearly onefold increase in paxillin expression was detected in hMSCs subjected to aligned nanofibers compared to those on randomly oriented nanofibers (Figure 6C). Our results may suggest that the enhancement in focal adhesion formation on aligned nanofibers could facilitate the osteogenesis potential of hMSCs.
Figure 6.
Effect of fiber topography on the focal adhesion formation of hMSCs.
Notes: (A) The location of paxillin in hMSCs cultured on glass. Scale bar indicates 20 μm. (B) Representative immunofluorescent images of paxillin expressed on different samples. Scale bars indicate 100 μm. DAPI (blue), phalloidin (green), and paxillin (red). (C) Western blot analysis of paxillin expression in hMSCs on different samples. GAPDH was used as control. *Compared with R-pDA, P<0.05; #compared with R-pDA-pep, P<0.05.
Abbreviations: hMSCs, human mesenchymal stem cells; DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; pDA, polydopamine; PCL, polycaprolactone; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide.
Conclusion
In this study, BFP-decorated PCL nanofibers with highly ordered structures that simulated the crucial characteristics of native bone tissue were developed via catechol chemistry to support and regulate osteoblast differentiation of hMSCs. Our results demonstrated that the surface modification of BFP-1 peptides on nanofibers could improve hMSCs adhesion, spreading, and proliferation. More importantly, the aligned nanofibers and surface-grafted BFP-1 peptides can act synergistically in combination to enhance the osteogenic differentiation of stem cells even under non-osteoinductive conditions. Other experiments are in progress to investigate whether the combination of osteoinductive peptides and aligned nanofibers could realize the synergistic effect on bone formation in vivo as well as in vitro. We believe the presented study provides an instructive insight in designing biomimetic scaffolds targeted at efficient osteogenic differentiation in the context of bone tissue engineering and regenerative medicine.
Supplementary materials
Quantification of immobilized peptides on the nanofibers
Immobilized peptides on the functionalized nanofibers were determined by fluorescamine assay. After surface modification, the supernatant was retrieved and reacted with fluorescamine solution (100 μg/mL in acetone; Aladdin Reagent Co, Ltd, Shanghai, People’s Republic of China) in a 96-well plate (bone-forming peptide solution =75 μL, fluorescamine solution =25 μL) for 5 minutes at room temperature. Then, the fluorescence intensities for each sample were measured at 365 nm excitation wavelength and 470 nm emission wavelength using a Multilabel Reader (PerkinElmer Inc., Waltham, MA, USA). The content of grafted peptides was calculated from a standard curve based on a series of peptide solutions with known concentrations. Moreover, the fluorescein isothiocyanate-labeled peptides were immobilized on the nanofibers to visualize the distribution and density of peptides on the fibers by laser scanning confocal microscope (Carl Zeiss Meditec AG, Jena, Germany).
LDH assay
The cytotoxicity of functionalized nanofibers was assessed using lactate dehydrogenase (LDH) assay kit (Beyotime Biotechnology, Shanghai, People’s Republic of China) following the manufacturer’s protocol. After 24 hours of culture, 10 μL of the supernatant was transferred to a 96-well plate along with 100 μL LDH reaction mix and incubated for 30 minutes at room temperature. Absorbance measurement was done at 450 nm. The supernatant of the untreated cells and the lysed cells on tissue culture plate (TCP) were used as negative (0%) and positive controls (100%), respectively. The LDH leakage (% of positive control) is expressed as the percentage of (ODtest − ODblank)/(ODpositive − ODblank), where ODblank is the optical density of supernatant from TCP group.
Visualization and determination of BFP-1 peptides immobilized on nanofibers.
Notes: The binding density of peptides on both randomly oriented and aligned nanofibers increased as the concentration of treated peptides rose (P<0.05). Though aligned fibers presented a little higher peptide density than randomly oriented fibers in each concentration group, but no statistical differences between them (P>0.05) were observed, suggesting the orientation has little effect on the grafting efficiency of peptides on the surface of nanofibers. Scale bars indicate 10 μm.
Abbreviation: BFP-1, bone-forming peptide-1.
The screening for optimal concentration of BFP-1 solution in surface functionalization.
Notes: (A) The cell proliferation of hMSCs cultured on aligned nanofibers treated with various concentrations of peptide for 1, 3, and 5 days. (B) ALP activity and (C) osteogenic gene expression of hMSCs cultured on aligned nanofibers treated with various concentrations of peptide at day 7. The results of ALP activity and early osteogenic genes (Runx2, ALP, and Col1a1) expression showed that the immobilized peptides on PCL nanofibers could enhance the osteogenic capacity of hMSCs in a concentration-dependent manner (0–200 μg/mL). However, high peptide concentration (400 μg/mL) on surfaces may inhibit osteogenic differentiation of hMSCs, which probably attribute to the cell growth restriction in high concentration group in accordance with CCK-8 results. Therefore, 200 μg/mL BFP-1 solution is an optimal concentration for surface functionalization of nanofibers. *P<0.05.
Abbreviations: hMSCs, human mesenchymal stem cells; ALP, alkaline phosphatase; Runx2, Runt-related transcription factor 2; Col1a1, Type 1 collagen alpha 1; PCL, polycaprolactone; CCK-8, Cell Counting Kit-8; BFP-1, bone-forming peptide-1; TCP, tissue culture plate; d, day; OCN, osteopontin.
SEM observation of adhering hMSCs on samples in serum-containing growth media.
Notes: Due to the effect of adhesive proteins, such as laminin and fibronectin, in serum, no evident difference in cell morphology was observed between R-pDA and R-pDA-pep groups as well as between A-pDA and A-pDA-pep groups at different time points. This finding was correlated with the result of subsequent focal adhesion formation assay, which revealed that there were no significant differences in the paxillin expression between R-pDA and R-pDA-pep groups as well as between A-pDA and A-pDA-pep groups when cells were cultured in the serum-containing media. The possible explanation is that abundant serum-derived adhesive proteins, such as laminin and fibronectin, were adsorbed on the surface of nanofibers, and enabled cells sufficiently spread out on all sample surfaces. Hence, the contribution of nonadhesive BFPs to cell adhesion and spreading was covered by the potent adhesive proteins. Scale bars indicate 15 μm.
Abbreviations: SEM, scanning electron microscope; hMSCs, human mesenchymal stem cells; pDA, polydopamine; BFP, bone-forming peptide; h, hours; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide; PCL, polycaprolactone.
Confocal micrographs of hMSCs on glass and nanofibers in serum-free growth media.
Notes: Cytoskeleton is colored green. Scale bars indicate 100 μm.
Abbreviations: hMSCs, human mesenchymal stem cells; pDA, polydopamine; h, hours; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide; PCL, polycaprolactone.
LDH release from hMSCs after culture for 24 hours on nanofibers (normalized to LDH release from positive control).
Notes: LDH is a stable enzyme present in the cytoplasm and released after cell death due to the compromise of the integrity of plasma membranes. Therefore, the relative value of LDH can serve as an indicator to reflect the proportion of dead cells in whole group. Approximately 8% dead cells were detected on the TCP surface at 24 hours after cell seeding. Compared to the control group, there was no considerable enhancement in LDH releasing level (P>0.05) when cells were exposed to nanofibers with different orientation and surface chemistries, suggesting favorable cytocompatability of functionalized nanofibers prepared in our work.
Abbreviations: LDH, lactate dehydrogenase; hMSCs, human mesenchymal stem cells; TCP, tissue culture plate; pDA, polydopamine; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide; PCL, polycaprolactone.
ALP and ARS staining of hMSCs on TCP.
Notes: Representative staining of ALP on TCP surface under non-osteoinductive condition (A) and osteoinductive condition (B) at day 14. ARS staining on TCP surface under non-osteoinductive condition (C) and osteoinductive condition (D) at day 21.
Abbreviations: ALP, alkaline phosphatase; TCP, tissue culture plate; ARS, Alizarin Red S; OM, osteoinductive media; α-MEM, α-Minimum Essential Medium; hMSC, human mesenchymal stem cell.
Table S1.
Primer sequences used for RT-PCR analysis
| Genes | 5′–3′ | Primers |
|---|---|---|
| Osteocalcin (OCN) | Forward | CCTGAAAGCCGATGTGGT |
| Reverse | AGGGCAGCGAGGTAGTGA | |
| Runt-related transcription factor 2 (Runx2) | Forward | AGGAATGCGCCCTAAATCACT |
| Reverse | ACCCAGAAGGCACAGACAGAAG | |
| Type I collagen alpha 1 (Col1a1) | Forward | AGACACTGGTGCTAAGGGAGAG |
| Reverse | GACCAGCAACACCATCTGCG | |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Forward | CGACAGTCAGCCGCATCTT |
| Reverse | CCAATACGACCAAATCCGTTG |
Abbreviation: RT-PCR, real-time polymerase chain reaction.
Table S2.
The roughness in different nanofiber-based platforms
| Specimen | Ra (μm, mean ± SD) | Sample | Ra (μm, mean ± SD) |
|---|---|---|---|
| R | 1.67±0.25 | A | 1.92±0.31 |
| R-pDA | 2.02±0.33 | A-pDA | 2.17±0.15 |
| R-pDA-pep | 2.74±0.42 | A-pDA-pep | 2.71±0.23 |
Abbreviations: SD, standard deviation; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pDA, polydopamine; PCL, polycaprolactone; pep, peptide.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (No 81371697, No 81271183), Peking University’s 985 Grants, the Open Project of Chongqing Key Laboratory of Oral Diseases and Biomedical Sciences (No ODBS-2014-001), Program for Innovation Team Building at Institutions of Higher Education in Chongqing in 2013, Chongqing Municipal Key Laboratory of Oral Biomedical Engineering of Higher Education, Chongqing Research Program of Basic Research and Frontier Technology (No cstc2013jcyjA10022) and the Outstanding Doctoral Scientific Research Fund of Chongqing Medical University. We would also like to thank Linqian Yang, Xucheng Xu, Xiaoli Ji, Shuai Zhang, Ting Ma, Mingyue Liu, and Yuying Shi for their help in this work.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gaharwar AK, Mihaila SM, Swami A, et al. Bioactive silicate nanoplatelets for osteogenic differentiation of human mesenchymal stem cells. Adv Mater. 2013;25(24):3329–3336. doi: 10.1002/adma.201300584. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Kim HN, Lim KT, et al. Synergistic effects of nanotopography and co-culture with endothelial cells on osteogenesis of mesenchymal stem cells. Biomaterials. 2013;34(30):7257–7268. doi: 10.1016/j.biomaterials.2013.06.029. [DOI] [PubMed] [Google Scholar]
- 3.Lim SH, Mao HQ. Electrospun scaffolds for stem cell engineering. Adv Drug Deliv Rev. 2009;61(12):1084–1096. doi: 10.1016/j.addr.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Yague MA, Abbah SA, McNamara L, Zeugolis DI, Pandit A, Biggs MJ. Biomimetic approaches in bone tissue engineering: integrating biological and physicomechanical strategies. Adv Drug Deliv Rev. 2015;84:1–29. doi: 10.1016/j.addr.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 5.Jang J-H, Castano O, Kim H-W. Electrospun materials as potential platforms for bone tissue engineering. Adv Drug Deliv Rev. 2009;61(12):1065–1083. doi: 10.1016/j.addr.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Yoo HS, Kim TG, Park TG. Surface-functionalized electrospun nanofibers for tissue engineering and drug delivery. Adv Drug Deliv Rev. 2009;61(12):1033–1042. doi: 10.1016/j.addr.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Cipitria A, Skelton A, Dargaville TR, Dalton PD, Hutmacher DW. Design, fabrication and characterization of PCL electrospun scaffolds – a review. J Mater Chem. 2011;21(26):9419–9453. [Google Scholar]
- 8.Lee J-H, Lee YJ, Cho H-J, Shin H. Guidance of in vitro migration of human mesenchymal stem cells and in vivo guided bone regeneration using aligned electrospun fibers. Tissue Eng Part A. 2014;20(15–16):2031–2042. doi: 10.1089/ten.tea.2013.0282. [DOI] [PubMed] [Google Scholar]
- 9.Ma J, He X, Jabbari E. Osteogenic differentiation of marrow stromal cells on random and aligned electrospun poly(l-lactide) nanofibers. Ann Biomed Eng. 2010;39(1):14–25. doi: 10.1007/s10439-010-0106-3. [DOI] [PubMed] [Google Scholar]
- 10.Nikkhah M, Edalat F, Manoucheri S, Khademhosseini A. Engineering microscale topographies to control the cell-substrate interface. Biomaterials. 2012;33(21):5230–5246. doi: 10.1016/j.biomaterials.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beachley V, Katsanevakis E, Zhang N, Wen X. Highly aligned polymer nanofiber structures: fabrication and applications in tissue engineering. Adv Polym Sci. 2011;246:171–212. [Google Scholar]
- 12.Kim HN, Jiao A, Hwang NS, Kim MS, Kim D-H, Suh K-Y. Nanotopography-guided tissue engineering and regenerative medicine. Adv Drug Deliv Rev. 2013;65(4):536–558. doi: 10.1016/j.addr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha C, Liechty WB, Khademhosseini A, Peppas NA. Designing biomaterials to direct stem cell fate. ACS Nano. 2012;6(11):9353–9358. doi: 10.1021/nn304773b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HK, Kim JH, Park DS, et al. Osteogenesis induced by a bone forming peptide from the prodomain region of BMP-7. Biomaterials. 2012;33(29):7057–7063. doi: 10.1016/j.biomaterials.2012.06.036. [DOI] [PubMed] [Google Scholar]
- 15.Perikamana SK, Lee J, Ahmad T, et al. Effects of immobilized BMP-2 and nanofiber morphology on in vitro osteogenic differentiation of hMSCs and in vivo collagen assembly of regenerated bone. ACS Appl Mater Interfaces. 2015;7(16):8798–8808. doi: 10.1021/acsami.5b01340. [DOI] [PubMed] [Google Scholar]
- 16.Marletta G, Ciapetti G, Satriano C, Pagani S, Baldini N. The effect of irradiation modification and RGD sequence adsorption on the response of human osteoblasts to polycaprolactone. Biomaterials. 2005;26(23):4793–4804. doi: 10.1016/j.biomaterials.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 17.Ko E, Yang K, Shin J, Cho SW. Polydopamine-assisted osteoinductive peptide immobilization of polymer scaffolds for enhanced bone regeneration by human adipose-derived stem cells. Biomacromolecules. 2013;14(9):3202–3213. doi: 10.1021/bm4008343. [DOI] [PubMed] [Google Scholar]
- 18.Rim NG, Kim SJ, Shin YM, et al. Mussel-inspired surface modification of poly(l-lactide) electrospun fibers for modulation of osteogenic differentiation of human mesenchymal stem cells. Colloids Surf B Biointerfaces. 2012;91:189–197. doi: 10.1016/j.colsurfb.2011.10.057. [DOI] [PubMed] [Google Scholar]
- 19.Kim MJ, Lee B, Yang K, et al. BMP-2 peptide-functionalized nanopatterned substrates for enhanced osteogenic differentiation of human mesenchymal stem cells. Biomaterials. 2013;34(30):7236–7246. doi: 10.1016/j.biomaterials.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Hollister SJ. Porous scaffold design for tissue engineering. Nat Mater. 2005;4(7):518–524. doi: 10.1038/nmat1421. [DOI] [PubMed] [Google Scholar]
- 21.Ku SH, Ryu J, Hong SK, Lee H, Park CB. General functionalization route for cell adhesion on non-wetting surfaces. Biomaterials. 2010;31(9):2535–2541. doi: 10.1016/j.biomaterials.2009.12.020. [DOI] [PubMed] [Google Scholar]
- 22.Low WC, Rujitanaroj PO, Lee DK, et al. Nanofibrous scaffold-mediated REST knockdown to enhance neuronal differentiation of stem cells. Biomaterials. 2013;34(14):3581–3590. doi: 10.1016/j.biomaterials.2013.01.093. [DOI] [PubMed] [Google Scholar]
- 23.Shin YM, Lee YB, Kim SJ, et al. Mussel-inspired immobilization of vascular endothelial growth factor (VEGF) for enhanced endothelialization of vascular grafts. Biomacromolecules. 2012;13(7):2020–2028. doi: 10.1021/bm300194b. [DOI] [PubMed] [Google Scholar]
- 24.Ku SH, Park CB. Combined effect of mussel-inspired surface modification and topographical cues on the behavior of skeletal myoblasts. Adv Healthc Mater. 2013;2(11):1445–1450. doi: 10.1002/adhm.201300067. [DOI] [PubMed] [Google Scholar]
- 25.Lee H, Dellatore SM, Miller WM, Messersmith PB. Mussel-inspired surface chemistry for multifunctional coatings. Science. 2007;318(5849):426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das RK, Zouani OF. A review of the effects of the cell environment physicochemical nanoarchitecture on stem cell commitment. Biomaterials. 2014;35(20):5278–5293. doi: 10.1016/j.biomaterials.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 27.Biggs MJP, Richards RG, Dalby MJ. Nanotopographical modification: a regulator of cellular function through focal adhesions. Nanomedicine. 2010;6(5):619–633. doi: 10.1016/j.nano.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang X, Cao HQ, Shi LY, Ng SY, Stanton LW, Chew SY. Nanofiber topography and sustained biochemical signaling enhance human mesenchymal stem cell neural commitment. Acta Biomater. 2012;8(3):1290–1302. doi: 10.1016/j.actbio.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 29.Kaur G, Wang C, Sun J, Wang Q. The synergistic effects of multivalent ligand display and nanotopography on osteogenic differentiation of rat bone marrow stem cells. Biomaterials. 2010;31(22):5813–5824. doi: 10.1016/j.biomaterials.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Kaur G, Valarmathi MT, Potts JD, Wang Q. The promotion of osteoblastic differentiation of rat bone marrow stromal cells by a polyvalent plant mosaic virus. Biomaterials. 2008;29(30):4074–4081. doi: 10.1016/j.biomaterials.2008.06.029. [DOI] [PubMed] [Google Scholar]
- 31.Kozhevnikova M, Mikaelyan A, Starostin V. Molecular and genetic regulation of osteogenic differentiation of mesenchymal stromal cells. Biol Bull. 2008;35(3):223–232. [PubMed] [Google Scholar]
- 32.Turner CE. Paxillin and focal adhesion signalling. Nat Cell Biol. 2000;2(12):231–236. doi: 10.1038/35046659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Visualization and determination of BFP-1 peptides immobilized on nanofibers.
Notes: The binding density of peptides on both randomly oriented and aligned nanofibers increased as the concentration of treated peptides rose (P<0.05). Though aligned fibers presented a little higher peptide density than randomly oriented fibers in each concentration group, but no statistical differences between them (P>0.05) were observed, suggesting the orientation has little effect on the grafting efficiency of peptides on the surface of nanofibers. Scale bars indicate 10 μm.
Abbreviation: BFP-1, bone-forming peptide-1.
The screening for optimal concentration of BFP-1 solution in surface functionalization.
Notes: (A) The cell proliferation of hMSCs cultured on aligned nanofibers treated with various concentrations of peptide for 1, 3, and 5 days. (B) ALP activity and (C) osteogenic gene expression of hMSCs cultured on aligned nanofibers treated with various concentrations of peptide at day 7. The results of ALP activity and early osteogenic genes (Runx2, ALP, and Col1a1) expression showed that the immobilized peptides on PCL nanofibers could enhance the osteogenic capacity of hMSCs in a concentration-dependent manner (0–200 μg/mL). However, high peptide concentration (400 μg/mL) on surfaces may inhibit osteogenic differentiation of hMSCs, which probably attribute to the cell growth restriction in high concentration group in accordance with CCK-8 results. Therefore, 200 μg/mL BFP-1 solution is an optimal concentration for surface functionalization of nanofibers. *P<0.05.
Abbreviations: hMSCs, human mesenchymal stem cells; ALP, alkaline phosphatase; Runx2, Runt-related transcription factor 2; Col1a1, Type 1 collagen alpha 1; PCL, polycaprolactone; CCK-8, Cell Counting Kit-8; BFP-1, bone-forming peptide-1; TCP, tissue culture plate; d, day; OCN, osteopontin.
SEM observation of adhering hMSCs on samples in serum-containing growth media.
Notes: Due to the effect of adhesive proteins, such as laminin and fibronectin, in serum, no evident difference in cell morphology was observed between R-pDA and R-pDA-pep groups as well as between A-pDA and A-pDA-pep groups at different time points. This finding was correlated with the result of subsequent focal adhesion formation assay, which revealed that there were no significant differences in the paxillin expression between R-pDA and R-pDA-pep groups as well as between A-pDA and A-pDA-pep groups when cells were cultured in the serum-containing media. The possible explanation is that abundant serum-derived adhesive proteins, such as laminin and fibronectin, were adsorbed on the surface of nanofibers, and enabled cells sufficiently spread out on all sample surfaces. Hence, the contribution of nonadhesive BFPs to cell adhesion and spreading was covered by the potent adhesive proteins. Scale bars indicate 15 μm.
Abbreviations: SEM, scanning electron microscope; hMSCs, human mesenchymal stem cells; pDA, polydopamine; BFP, bone-forming peptide; h, hours; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide; PCL, polycaprolactone.
Confocal micrographs of hMSCs on glass and nanofibers in serum-free growth media.
Notes: Cytoskeleton is colored green. Scale bars indicate 100 μm.
Abbreviations: hMSCs, human mesenchymal stem cells; pDA, polydopamine; h, hours; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide; PCL, polycaprolactone.
LDH release from hMSCs after culture for 24 hours on nanofibers (normalized to LDH release from positive control).
Notes: LDH is a stable enzyme present in the cytoplasm and released after cell death due to the compromise of the integrity of plasma membranes. Therefore, the relative value of LDH can serve as an indicator to reflect the proportion of dead cells in whole group. Approximately 8% dead cells were detected on the TCP surface at 24 hours after cell seeding. Compared to the control group, there was no considerable enhancement in LDH releasing level (P>0.05) when cells were exposed to nanofibers with different orientation and surface chemistries, suggesting favorable cytocompatability of functionalized nanofibers prepared in our work.
Abbreviations: LDH, lactate dehydrogenase; hMSCs, human mesenchymal stem cells; TCP, tissue culture plate; pDA, polydopamine; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pep, peptide; PCL, polycaprolactone.
ALP and ARS staining of hMSCs on TCP.
Notes: Representative staining of ALP on TCP surface under non-osteoinductive condition (A) and osteoinductive condition (B) at day 14. ARS staining on TCP surface under non-osteoinductive condition (C) and osteoinductive condition (D) at day 21.
Abbreviations: ALP, alkaline phosphatase; TCP, tissue culture plate; ARS, Alizarin Red S; OM, osteoinductive media; α-MEM, α-Minimum Essential Medium; hMSC, human mesenchymal stem cell.
Table S1.
Primer sequences used for RT-PCR analysis
| Genes | 5′–3′ | Primers |
|---|---|---|
| Osteocalcin (OCN) | Forward | CCTGAAAGCCGATGTGGT |
| Reverse | AGGGCAGCGAGGTAGTGA | |
| Runt-related transcription factor 2 (Runx2) | Forward | AGGAATGCGCCCTAAATCACT |
| Reverse | ACCCAGAAGGCACAGACAGAAG | |
| Type I collagen alpha 1 (Col1a1) | Forward | AGACACTGGTGCTAAGGGAGAG |
| Reverse | GACCAGCAACACCATCTGCG | |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Forward | CGACAGTCAGCCGCATCTT |
| Reverse | CCAATACGACCAAATCCGTTG |
Abbreviation: RT-PCR, real-time polymerase chain reaction.
Table S2.
The roughness in different nanofiber-based platforms
| Specimen | Ra (μm, mean ± SD) | Sample | Ra (μm, mean ± SD) |
|---|---|---|---|
| R | 1.67±0.25 | A | 1.92±0.31 |
| R-pDA | 2.02±0.33 | A-pDA | 2.17±0.15 |
| R-pDA-pep | 2.74±0.42 | A-pDA-pep | 2.71±0.23 |
Abbreviations: SD, standard deviation; R, pure randomly oriented PCL nanofiber; A, pure aligned PCL nanofiber; pDA, polydopamine; PCL, polycaprolactone; pep, peptide.