Table 2.
Top prescription drugs predicted to target DAO
| Drug | DAO SEA E valuea | Affinity binb (μM) | Max Tcc | IC50 from literatured | |
|---|---|---|---|---|---|
| 1 |
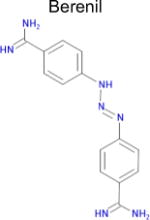
|
4.4 × 10−30 (rat)e | 10 | 0.36 Tc | 13 ± 1f nM |
| 2 |
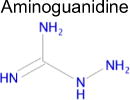
|
1.8 × 10−19 | 1 | 1.00 Tc (known in ChEMBL) | 10–1508 nM |
| 3 |
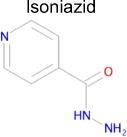
|
1.1 × 10−16e | 1 | 0.28 Tc | 970± 50f μM |
| 4 |
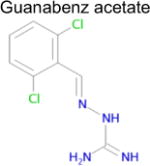
|
6.8 × 10−14 (rat) | 10 | 0.38 Tc | 5.1 ± 0.8g μM |
| 5 |
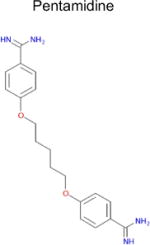
|
2.2 × 10−8 (rat)e | 10 | 0.29 Tc | 0.29–3f μM |
| 6 |
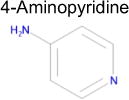
|
4.4 × 10−5 | 1 | 0.36 Tc | Not known in literature |
| 7 |

|
7.4 × 10−5 | 1 | 0.35 Tc | Not known in literature |
| 8 |
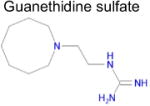
|
3.5 × 10−4 | 1 | 0.32 Tc | Not known in literature |
| 9 |
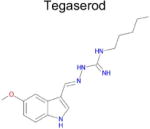
|
4.8 × 10−4 (rat) | 10 | 0.44 Tc | Not known in literature |
| 10 |
|
(Not predicted) | Not predicted and thus no value | Not predicted and thus no value | 90 ± 14f μM |
| 11 |

|
(Not predicted) | Not predicted and thus no value | Not predicted and thus no value | 100 ± 8g μM |
The descriptions of the data shown in this table is described below
E-value is derived from a statistical model which presents the probability of observing this raw score by random chance alone. The model is similar to that underlying BLAST [45]. The smaller the E-value, the stronger it is, and this indicates strong overall chemical structural similarity between two sets of compounds. Typically with small E-values, <0.001, are unlikely to be random chance [74]
Affinity bin The ligands for each protein target that is used as a reference (e.g., DAO) are subdivided (“binned”) by the logs of their affinities, and a SEA calculation is run independently against each bin. Thus, when asking whether a particular drug has a strong SEA E-value for DAO, we first compute the E-value for the compound against DAO ligands at 0.1 μM, DAO ligands at 1 μM,…, DAO ligands at 10 μM, and finally report the affinity bin at which DAO achieves its strongest E-value for the drug of interest. This may be thought of as an approximate prediction of the drug’s binding affinity at the target, but this interpretation has not been tested at scale
Tc Tanimoto coefficient. The Tc calculates the number of “on” bits in common between two fingerprints, divided by the total number of non-overlapping “on” bits between them. It is an overall measure of the similarity between any two molecular fingerprints, on a 0.0 (completely dissimilar) to 1.0 (completely similar) scale
IC50 IC50 value indicates the concentration needed to inhibit a biological function by half (by 50 %). In this study, it will be the concentration needed to inhibit formation of the product by diamine oxidase using putrescine as the substrate
Predicted using ChEMBL-14 (p-value) instead of ChEMBL-16 (E-value)
IC50 values (inhibition concentrations of DAO by 50 %) were obtained from literature [75]
