Abstract
Low energy electrons (LEEs) of energies less than 20 eV are generated in large quantities by ionizing radiation in biological matter. While LEEs are known to induce single (SSBs) and double strand breaks (DSBs) in DNA, their ability to inactivate cells by inducing nonreparable lethal damage has not yet been demonstrated. Here we observe the effect of LEEs on the functionality of DNA, by measuring the efficiency of transforming Escherichia coli with a [pGEM-3Zf (–)] plasmid irradiated with 10 eV electrons. Highly ordered DNA films were prepared on pyrolitic graphite by molecular self-assembly using 1,3-diaminopropane ions (Dap2+). The uniformity of these films permits the inactivation of approximately 50% of the plasmids compared to <10% using previous methods, which is sufficient for the subsequent determination of their functionality. Upon LEE irradiation, the fraction of functional plasmids decreased exponentially with increasing electron fluence, while LEE-induced isolated base damage, frank DSB, and non DSB-cluster damage increased linearly with fluence. While DSBs can be toxic, their levels were too low to explain the loss of plasmid functionality observed upon LEE irradiation. Similarly, non-DSB cluster damage, revealed by transforming cluster damage into DSBs by digestion with repair enzymes, also occurred relatively infrequently. The exact nature of the lethal damage remains unknown, but it is probably a form of compact cluster damage in which the lesions are too close to be revealed by purified repair enzymes. In addition, this damage is either not repaired or is misrepaired by E. coli, since it results in plasmid inactivation, when they contain an average of three lesions. Comparison with previous results from a similar experiment performed with γ-irradiated plasmids indicates that the type of clustered DNA lesions, created directly on cellular DNA by LEEs, may be more difficult to repair than those produced by other species from radiolysis.
INTRODUCTION
Secondary electrons are the most abundant secondary species generated by the transfer of energy from ionizing radiation (IR) into a condensed medium.1–5 Most of these consist of electrons with initial kinetic energies below 30 eV. A detailed knowledge of the reactions of such low-energy electrons (LEEs) with DNA is thus essential to understand and accurately describe the damage induced in DNA by ionizing radiation. LEEs have been shown to induce DNA damage such as single-strand breaks (SSBs) and frank double-strand breaks (DSBs)2,6–8 as well as damage to DNA bases,6,8–10 deoxyribose,6,8,11 and the phosphate group.6,9,10 LEEs with energies below 15 eV induce DNA fragmentation mainly via the formation of transient negative ions (TNIs), decaying into dissociative electron attachment (DEA) and autoionization, which produces dissociative electronically excited states.6
SSBs and isolated base DNA damage, which occur much more frequently than DSBs, can be repaired rapidly with high fidelity.12 It is generally accepted that DNA DSBs are the most frequent toxic DNA lesion induced by IR.13,14 A DSB is a type of clustered DNA damage in which two SSBs are formed in close proximity on opposite DNA strands, by either a one or two hit damage process or during DNA repair. Large-scale deletions and cell death can be induced by unrepaired DSBs.15,16 Moreover, misrepaired DSBs can give rise to mutations and chromosomal rearrangements, and hence cell death or cancer in multicellular organisms. A single unrepaired DSB in the DNA of Escherichia coli is lethal.17 DSBs can also be generated by enzymatic misrepair of other lesions in DNA such as SSBs and base damage.18–20
Recent results show that IR exposure of DNA in solution, or within human cells at low irradiation levels, induces bistranded clustered DNA damage, such as DNA oxidized sites (i.e., two or more oxidized purines, oxidized pyrimidines, abasic sites, or strand breaks within a few helical turns).12,21 In irradiated DNA, DSBs account for about 20% of the total of such complex damage, and the remaining 80% is non-DSB clusters.22–26 IR can also induce cross-links between DNA and nearby biomolecules, including proteins, which can cause cell death, mutagenesis or carcinogenesis.27 If not correctly repaired, the formation of DNA–protein cross-links within the cell may disrupt DNA transcription, replication, and, ultimately, cell division. It has been shown that the nucleosomal core histones (H2A, H2B, H3 and H4) are the main proteins involved in the formation of DNA–protein cross-links in irradiated chromatin.27
Although DSBs and other cluster lesions, as well as DNA–protein cross-links have the potential to be lethal, the relative contribution of each lesion to the loss of DNA functionality induced by LEEs in living cells is not known. Since LEEs have a very short-range in condensed matter (5–10 nm),3 it is not possible to irradiate the DNA of living cells with them. Moreover, in most of the previous experiments,6 suitable thin films of extracellular DNA for irradiation by LEEs were prepared by freeze-drying (lyophilization). Unfortunately, this method restricts damage to only a small fraction (<10%) of the DNAs in the film;2,6,28 such films have a highly irregular thickness, and thus more susceptible to charging29,30 such that LEEs are increasingly repelled as charging increases. Thus, a number of DNA targets cannot be irradiated,31 as this phenomenon limits the fraction of plasmids that can be damaged by LEEs. Both intra- and intermolecular electron stabilization and DEA result in film charging.32,33 These two phenomena are the main obstacles for the study of the effects of LEEs on DNA functionality, which requires a much larger percentage of damage than obtained with lyophilized films.
To overcome this problem, we constructed DNA films with a molecular, self-assembly technique, by which 1,3-diaminopropane molecules (Dap2+) are intercalated with plasmid DNA on a pyrolitic graphite substrate.34 Such films have a high uniformity compared to those prepared by lyophilization and more than 50% of plasmids can be irradiated by LEEs in a 5 monolayer film (ML).34 Furthermore, the binding of Dap2+ to DNA mimics that of the amino groups of the lysine and arginine amino acids within the basic, positively charged histone proteins (Scheme 1).
Scheme 1.
In vivo, positively charged polyamines also interact with the negatively charged DNA.35 Polyamines are believed to stabilize chromatin by compacting DNA and neutralizing negative charges.36 Diamines such as putrescine (Scheme 1) protect DNA against ionizing radiation either by inducing DNA compaction and aggregation or by acting as scavengers of free radicals, predominantly of the •OH type.36,37 However, recently Iacomino et al. demonstrated that polyamines were ineffective in conferring DNA protection at sub-mM concentrations.38
Owing to this advance in sample preparation, we present herein a novel method to analyze LEE-induced damage to DNA under conditions found in living cells. We measure cell survival as a function of LEE-induced damage to the pGEM-Zf(–) plasmid (i.e., supercoiled) DNA, which possesses an ampicillin resistance gene. The plasmid is bombarded in ultrahigh vacuum (UHV) with 10 eV electrons. This energy is typical of that of secondary electrons generated in condensed organic matter by ionizing radiation; it is close to the most probable initial kinetic energy of a secondary electron immediately after ionization, as calculated by Monte Carlo simulations.3 Moreover, previous measurements of strand-breaks induced in plasmid DNA by 0.5–20 eV electrons2,6 indicate the presence of a maximum in the yields, due to the formation of TNIs, around 10 eV. After bombardment, one portion of the sample serves for the analysis of various damages, including clustered lesions. Another portion is introduced into E. coli JM109 bacteria, which are then incubated with antibiotic ampicillin. These bacteria are devoid of any genes coding for resistance to ampicillin and thus rely on the integrity of pGEM-Zf(–) plasmid for survival. In other words, lethal damage to the plasmids reduces cellular transformation efficiency. We measure previously unreported LEE-induced DNA lesions, including base damage and clustered lesions and show how these new lesions and those previously detected influence cellular toxicity (i.e., the loss of DNA functionality or “viability” in bacterial cells). We find that the transformation efficiency decreases with increasing electron fluence, but only a small proportion of this decrease can be attributed to the formation of DSBs. Moreover, when damaged DNA is treated with DNA glycosylases, to convert base damage and apurinic/apyrimidinic (AP) sites to DSBs, non-DSB cluster damage can only account for a small fraction of observed loss of DNA functionality. Thus, other toxic, potentially lethal lesions, formed at much higher frequencies than DSBs and non-DSB cluster damage must be responsible for the observed loss of DNA functionality.
MATERIALS AND METHODS
Many of the experimental techniques used for the present studies have been described separately in detail elsewhere.28,39–42 Here we provide brief descriptions of each.
Plasmid DNA Preparation
Plasmid DNA [pGEM-3Zf(–), 3197 base pairs, ca.1 968 966 amu per plasmid] was extracted from E. coli JM109 and purified with a HiSpeed plasmid Maxi kit (QIAGEN) as previously described.28,31,40
Sample Preparation and LEE Irradiation
DNA was deposited onto freshly cleaved graphite (HOPG, ZYA grade, NT-MDT) by a soft adsorption method.34 The solution of DNA at a concentration of 200 ng/μL was mixed with a solution containing protonated 1,3-diaminopropane molecules (Dap2+) at a molar ratio of R[[Dap2+]/[PO4–]DNA] = 16. This solution was mixed and incubated at 25 °C for 15 min in the presence of TE buffer (pH = 8).34 The soft adsorption method was used for DNA deposition onto graphite substrate.34 A 50 μL droplet of DNA–Dap mixture was deposited onto a fresh HOPG surface and incubated for 15 min. The surplus of mixture solution was removed using filter paper. The average thickness of the film was 5 ML of DNA–Dap complexes.34 Afterward, the Dap–DNA films were placed on sample holders inside an UHV chamber equipped with an electron irradiator. The latter consists of an FRA-2X1-2 electron gun from Kimball Physics producing a beam adjustable in energy between 5 and 1000 eV. The spot size of the beam can be varied between 2 and 50 mm diameter at working distances between 10 and 50 mm. In the present experiment, it was set to irradiate an area of about 1.0 cm2, which was 3.5 times larger than the DNA sample. The chamber was evacuated for 24 h by a hydrocarbon-free turbo molecular pump to a base pressure of 5 × 10−9 Torr at room temperature. After stabilization of the electron-beam current at 3 nA (±5%), corresponding to the current density of 1.5 × 1011 electrons s−1 cm−2, the DNA films were individually irradiated with electrons of 10 eV for periods between 0 and 50 min at room temperature. While a sample was irradiated, the others were shielded from stray electrons by applying on them a repulsive potential of 9 V, with respect to the cathode of the electron gun. One sample placed in the UHV chamber was never irradiated with electrons to serve as a control. After irradiation, the samples were removed from the chamber and immediately dissolved in TE buffer at pH 8.0.
Transformation by LEE Irradiated Plasmid DNA
The transformation efficiency of the control and irradiated plasmids was determined using competent E. coli JM109. The suspension of competent cells was divided into 100 μL portions and each culture was mixed with 1 μL of either nonirradiated or irradiated samples, and then placed on ice for 30 min. After heat shock for 45 s at 42 °C, 900 μL Super Optimal Broth (SOC) was immediately added. After a shaking period of 1 h at 37 °C, the samples were spread on (LB) agar plates containing ampicillin at a concentration 50 μg/μL43 to select transformants (i.e., colonies which can grow in the presence of ampicillin). After the incubation at 37 °C overnight, the transformants were counted. The efficiency of the transformation is reflected by the number of transformants per microgram of DNA. All transformation experiments were repeated three times.
Transformation Efficiency Modeling
A supercoiled DNA plasmid is converted into circular and linear forms due to the induction of SSB and DSB, respectively, by IR.44 The efficiency of each of these DNA forms to transform bacteria has been shown to differ, but the literature is controversial; some studies suggested that supercoiled (S) and circular (C) DNA has the same probability to transform bacteria; however, others propose that S DNA has a higher probability to cross bacteria membrane and transform bacteria.43,45–47 The main parameters that determine transformation efficiency are the probabilities that the supercoiled, circular or linear forms (ρS, ρC and ρL, respectively) of plasmid DNA cross the cytoplasmic membrane (i.e., entry function) and the proportion of plasmids that remains capable of replicating and expressing the ampicillin resistance gene (i.e., survival function). Mathematically, the transformation efficiency can be described by
| (1) |
where the first and second bracketed terms are entry and survival function, respectively.42 S, C, and L denote the relative percentage of the supercoiled, circular, and linear forms of DNA, respectively, and φ is the electron fluence (in electron per cm2). βR is an effective cross section (in cm2 per electron) and it represents the yield of lethal damage; then, the product βRφ corresponds to the mean number of toxic lesions induced per plasmid. βR is independent of DNA conformation. The integer n is the number of toxic lesions needed to inactivate the plasmid.
Agarose Gel Electrophoresis and Image Analysis
The different topological forms of DNA were separated by 1% agarose gel in 1× TAE buffer (40 mM tris acetate and 1 mM EDTA at pH 8.0) at 100 V for 7 min and 75 V for 68 min (5 V· cm−1).48 SYBR Green I prestained both gel and DNA samples (Molecular probes) at a concentration of 10 000× and 100×, respectively. After electrophoresis, gels were scanned with a Typhoon-Trio laser scanner (from GE Healthcare) adjusted for the blue fluorescent mode at an excitation wavelength of 488 nm and filter type 520 nm band-pass (520 BP 40) in the normal sensitivity mode.48 The percentage of each form was obtained from Image Quant 5.0 (Molecular Dynamics) software analysis. These values were corrected for the weaker binding of SYBR Green I to the supercoiled form of DNA relative to those for nicked circular and linear configurations.49 To determine the correction factor, the same amounts (70 ng) of supercoiled and linear DNA were run in gel electrophoresis. After measuring the intensity of each band, the area under the linear DNA peak was divided by the area under the peak of supercoiled DNA to give the correction factor. In the present study, the correction factor was 1.5, which was employed to the quantification of plasmid DNA. Equations 2, 3 and 4 give the corrected amounts of S, C, and L in DNA samples:
| (2) |
| (3) |
| (4) |
where SG, CG, or LG are the integral values (i.e., the area under the peak) of the bands corresponding to the supercoiled, circular, and linear forms of DNA, respectively.
Detection of Enzyme-Sensitive DNA Damage
Each analyzed sample was split into two equal volumes, the first one for treatment with E. coli base excision repair endonuclease III (Nth) and/or formamidopyrimidine-DNA N-glycosylase (Fpg) (Trevigen, Inc.) and the second for mock treatment without enzyme. Nth and Fpg act both as N-glycosylases and AP-lyases. Nth can recognize and remove several damaged pyrimidines including 5,6 dihydroxythymine, thymine glycol, 5-hydroxy-5 methylhydanton, uracil glycol, 6-hydroxy-5, 6-dihdrothimine and methyltartronylurea.50 Fpg recognizes and removes several damaged purines including: 7,8-dihydro-8-oxoguanine (8-oxoguanine), 8-oxoadenine, fapy-guanine, methyl-fapy-guanine, fapy-adenine, aflatoxin B1-fapy-guanine, 5-hydroxy-cytosine and 5-hydroxy-uracil.51 The samples were categorized into three groups depending upon the postirradiation treatment. The first group termed frank SSBs or frank DSBs was maintained at –20 °C, the second frank + heat labile site SSB was incubated at 37 °C for 60 min in the absence of enzyme and the third (100 ng) was mixed with one unit of Nth or Fpg or a mixture of both and incubated at 37 °C for 60 min (1 unit of enzyme is defined as resulting in the cleavage at an AP site of an oligonucleotide at a rate of 1 pmol/h). Control samples were also incubated in the presence of incubation buffer only. Additionally, we tested the effect of increasing the amount of enzyme and incubation time on the additional yields of breaks produced by Fpg and Nth. Additional enzyme (20 U) was added after 60 min of incubation to ensure that all sensitive sites had been detected (data are not shown). At 1 U for both enzymes (Nth and Fpg), saturation occurred and no significant effect of additional Nth and Fpg incubation on the background levels of damage was observed for all the conditions studied. The enzyme reactions were quenched by addition of 10 μL of 0.5 M EDTA buffer and then mixed with loading buffer and kept on ice prior to electrophoresis.52 The yields of SSBs and DSBs were measured by gel electrophoresis.
Yields for the Formation of Specific Lesions in Irradiated Plasmid DNA
The yield of a given damage is expressed as a percentage of the total amount of DNA in the target per unit of fluence. The fluence-response curve for the formation of SSBs, as a fraction of intact supercoiled DNA, and loss of the supercoiled configuration were plotted as a linear and logarithmic function of incident electron surface density (i.e., the fluence). The yields of SSBs were obtained from the slope at zero fluence of the logarithmic curve fitted to the data.53,54 The yield of DSBs was determined from the slope of the linear electron fluence-response curve of the fractional abundance of the linear form of DNA.55 For the isolated base lesions, the yields were calculated by subtraction of the yield of plasmids with SSBs that had received only mock enzyme treatment from the yield of plasmids with SSBs following enzyme treatment.56 The yields of non-DSBs clustered damage were determined from the difference in the yields of DSBs determined in the presence and absence of enzymatic treatment of irradiated DNA.56
DNA Damage Modeling
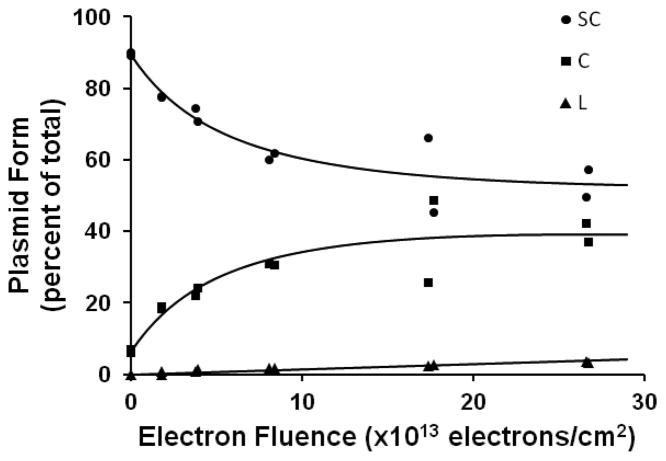
Before using eq 1 for modeling the results of the transformation, we must first choose algebraic functions S(φ), C(φ), and L(φ), which reproduce the gel electrophoresis data for the dependence of the different plasmid conformations on the electron fluence (Figure 1).
Figure 1.
Agarose gel electrophoresis of 10 eV-electrons irradiated extracellular plasmid pGEM-3Zf (–). Loss of supercoiled DNA (●) and the formation of circular (■) and linear (▲) DNA as a function of electron fluence.
For this purpose, we used the model described by Rezaee et al.31 to fit the loss of supercoiled plasmids. Originally, this model was designed to account for the low penetration of LEEs in condensed phase macromolecular samples such as DNA. Additionally, it introduces a term to account for the charging of the sample at long bombardment times. In our case, the algebraic expression to model the percentage of supercoiled plasmid as a function of electron fluence takes the form:
| (5) |
Where S0 is the initial percentage of supercoiled DNA before irradiation, h is the thickness of the sample and λ is the characteristic attenuation length of the 10 eV electron beam; ϕ is the “charging fluence constant” that can be seen as the characteristic fluence showing charging effects. Y is the total yield of SSBs and DSBs. We will not discuss here the values of these parameters, since our sole aim is to produce a function, eq 5, which fits the gel electrophoresis data (Figure 1). The growth of the linear form L(φ) is modeled by a straight line, where the initial value L = L0 is set to zero (eq 6). The evolution of circular form C(φ) with respect to the electron fluence is then derived from the difference between the sum of the initial values S0 and C0, and the sum of the functions S(φ) and L(φ):
| (6) |
| (7) |
RESULTS
DNA Strand Breaks Induced by 10 eV Electrons
Figure 1 shows the electron fluence-response curves for the formation of the supercoiled, nicked circle and linear forms in LEE-irradiated DNA followed by laser scanning of the bands in agarose gel. As expected, the fraction of supercoiled DNA in the sample decreased with increasing electron fluence and reached a plateau at approximately 52% supercoiled concentration; the fraction of both circular and linear DNA increased, with the circular form reaching a maximum at 39%, whereas linear DNA increased in a linear fashion with no plateau.
Transformation Efficiency of DNA Irradiated with LEEs
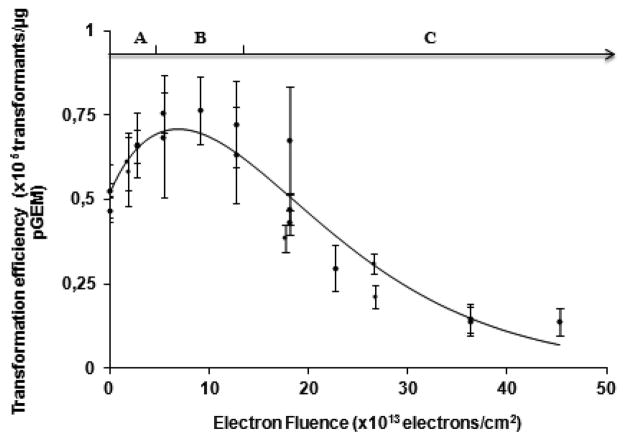
Figure 2 shows the transformation efficiency of DNA as a function of incident electron fluence in electrons·cm−2. The curves are fitted to the measured data using eq 1. The transformation efficiency curve can be divided in three regions A, B and C (Figure 2). The A region extends from zero to 5 × 1013 electrons/cm2 and is highly dependent on the ρc/ρs parameter, (i.e., the ratio of probabilities of circular and supercoiled plasmids to enter the bacteria), since in this dose range supercoiled DNA is converted to circular DNA. The B region ranges from 5 to 15 × 1013 electrons/cm2 and is highly influenced by the n parameter. Finally, the C region from 15 to 45 × 1013 electrons/cm2 reflects the magnitude of the yield of lethal damage (βR).
Figure 2.
Impact of 10 eV-electron irradiation on transformation efficiency. The continuous line is calculated with eq 1. The best fit to the data occurs when the ratio of ρC/ρS = 3, ρL = 0, n = 3 and βR = (1.2 ± 0.1) × 10−14 cm2 per electron. Each data point represents the mean of three independent measurements, obtained on the same day. At certain fluences, more than one data point was recorded. The error bars correspond to the standard deviation for each group of three measurements.
The best fit was obtained at ρc/ρs = 3, which reflects the fact that the efficiency of circular DNA to enter the bacteria (ρc) is greater than for supercoiled DNA (ρs), by a factor of 3, as previously reported.42 As well, the best fit was obtained at n = 3, indicating that, on average, three toxic lesions are required to inactivate the plasmid. Finally, for 10 eV electrons the yield for the production of a lethal lesion is given by βR = (1.2 ± 0.1) × 10−14 cm2/electron, where βR represents the yield of lethal damage. The presence of 1,3-diaminopropane molecules had no effect on the transformation efficiency at zero fluence.
Effect of LEEs on the Biological Activity of Plasmid DNA
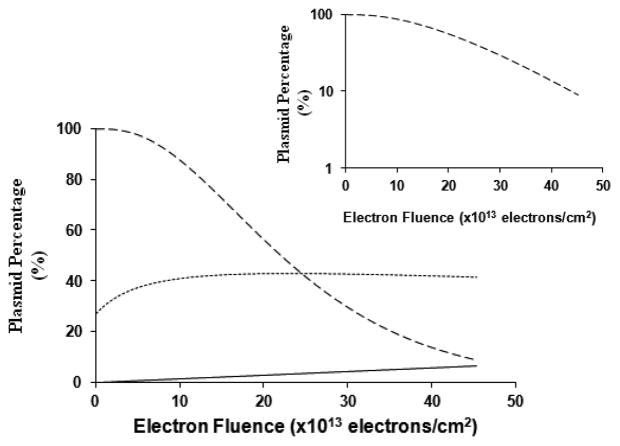
A functional plasmid (pGEM-3Zf-) results in the formation of a bacterial colony, indicating that it replicates and also expresses the ampicillin resistance gene. Figure 3 shows the entry efficiency and functionality (i.e., the two components of the transformation efficiency) of the plasmid transformed into wild type E. coli (JM 109) as a function of electron fluence. These curves were calculated from eq 1 with the parameters fitted to produce the curve of Figure 2.
Figure 3.
Biological activity of plasmid pGEM-3Zf (–) following 10 eV-electron irradiation. The “survival function” (dashed curve) of the plasmid is calculated using eq 1. The dotted line represents the variation in the “entry function” as a function of electron fluence (eq 1). The behavior is due to the change in the number of plasmids with different entry coefficients. For comparison, the formation of linear plasmids as a function of electron fluence is also shown (straight line). The inset shows the functionality of plasmid DNA on a logarithmic scale.
The survival curve in Figure 3 (plasmid functionality) has a definite plateau at low electron fluence followed by an exponential loss of viability. This behavior suggests that the lethal lesions can be repaired, when present at low levels, but at higher levels the repair process is overwhelmed and results in plasmid inactivation. At a fluence of 20 × 1013 electrons/cm2, 56% of the plasmids are functional, while at 40 × 1013 electrons/cm2, this percentage is reduced to 13%.
Isolated Base Damage Induced by LEEs in Plasmid DNA
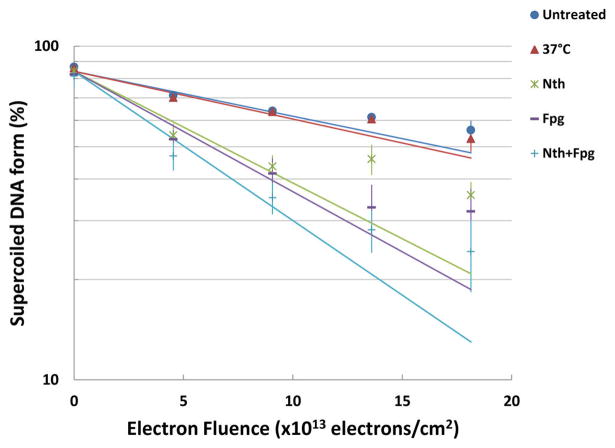
A semilogarithmic plot of the loss of the supercoiled configuration with increasing electron fluence and treatment with DNA glycosylases is shown in Figure 4.
Figure 4.
Loss of supercoiled pGEM-3Zf (–) DNA versus electron fluence. After irradiation, DNA was untreated (closed circles) or incubated at 37 °C (closed triangles) or incubated with Nth (asterisk) or Fpg (dashes) or both enzymes Nth and Fpg (pluses). Linear fits obtained from data recorded at electron fluences < 1012 electrons/cm2.
Post-treatment of irradiated DNA at 37 °C with either Nth or Fpg, which act both as N-glycosylases and AP-lyases, led to a further decrease in the supercoiled form of the plasmid. The yields for the formation of both Nth and Fpg sensitive sites (i.e., oxidized pyrimidine and purine bases, respectively), determined from the fluence–response curves for the loss of supercoiled DNA, are shown in Table 1. The net yields for the formation of enzyme-sensitive sites, given in Table 1, were obtained57 with the procedures described in the section Materials and Methods of ref 42. The yield for purine base damage in DNA was YFpg-SS = (50.0 ± 0.8) × 10−16 cm2/electron vs pyrimidine base damage yield YNth-SS = (44.0 ± 0.7) × 10−16 cm2/electron. The yield of total base damage was Y(Fpg+Nth)-SS = (70.0 ± 1) × 10−16 cm2 per electron.
Table 1.
Yields (Y × 10−16 cm2/electron) of SSB in pGEM-3-Zf (–) Plasmid DNA Induced by 10 eV-Electron Irradiation Followed by Incubation with Nth and Fpga
| YF(SSB) | YHSS | YFpg-SS | YNth-SS | Y(Fpg+Nth)-SS |
|---|---|---|---|---|
| 31 ± 5 | 2.0 ± 0.3 | 50 ± 8 | 44 ± 7 | 70 ± 10 |
YF(SSB), YHLS, YFpg-SS, YNth-SS and Y(Fpg + Nth)-SS are the yields of SSBs without enzymatic treatment and followed by heating at 37 °C, from purine and pyrimidine base damage and total base damage, respectively.
Clustered Base Damage Induced by LEEs in Plasmid DNA
Figure 5 shows fluence-response curves for the formation of linear plasmid DNA, under conditions identical to those, which produced the data of Figure 4.
Figure 5.
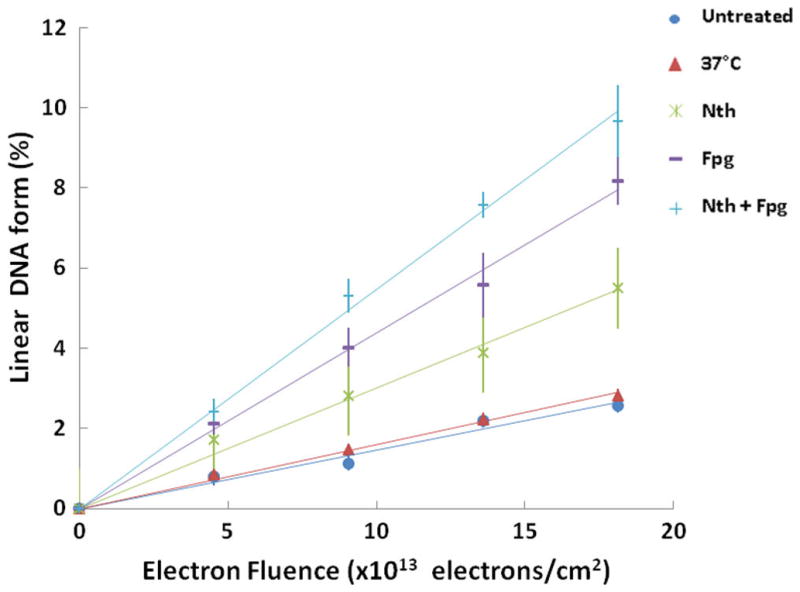
Fluence–response plots showing the fractions of linear DNA. After irradiation, DNA was untreated (close circles) or incubated at 37 °C (close triangles) or incubated with Nth (asterisk) or Fpg (dashes) or both enzymes Nth and Fpg (pluses).
Incubation of the irradiated plasmid DNA with either Nth or Fpg at 37 °C results in the formation of linear DNA, when the breaks induced by these enzymes at the sites of damaged bases are either in close proximity to one another or to a SSB, but on opposite strands. The respective curves for total DSB yields (i.e., Nth-induced DSBs + Frank DSBs as well as Fpg-induced DSBs + Frank DSBs), as shown in Figure 5, increase linearly with radiation dose. The levels of clustered DNA damage revealed by Nth and Fpg greatly exceed the number of frank DSBs. Table 2 presents the yield for enzyme-sensitive sites detected as additional DSBs. It shows that the yield of total non-DSB cluster damage was about 2.6 times higher than that of frank DSBs (3.9 ± 0.6) × 10−16 cm2/electron vs (1.5 ± 0.2) × 10−16 cm2/electron.
Table 2.
Yields (Y × 10−16 cm2/electron) of DSB Induction in pGEM-3-Zf (–) Plasmid DNA by 10 eV-Electron Irradiation with No Further Treatment and Followed by Heating to 37 °C and Incubation with Nth, Fpg, and Botha
| YF(DSB) | Y HLS | YFpg-DS | YNth-DS | Y(Fpg+Nth)-DS |
|---|---|---|---|---|
| 1.5 ± 0.2 | 0.14 ± 0.02 | 2.8 ± 0.4 | 1.4 ± 0.2 | 3.9 ± 0.6 |
The designations are the same as in Table 1 with SSBs being replaced by DSBs.
DISCUSSION
The dashed survival curve in Figure 3, which represents the functionality of irradiated pGEM-3Zf (–) plasmid, has an initial plateau (see also inset) followed by a decrease at higher electron fluence. The initial plateau suggests that the lethal lesions can be repaired when present at low levels, as proposed for other types of genotoxic damage.58,59 At higher levels of damage, the repair process is overwhelmed and results in plasmid inactivation. As seen from Figure 1, beyond 1.5 × 1014 electrons/cm2, the small fraction of plasmids left in the supercoiled configuration remains essentially constant. This “saturation”, which results from the exponential decay of the initial target (i.e., supercoiled DNA),31 can be correlated with the near-linear decrease of functionality shown in Figures 2 and 3. The constant percentage of supercoiled and circular DNA, observed beyond 1.5 × 1014 electrons/cm2, can be attributed to film charging, which repels incoming electrons. Charging occurs because the electrons attenuation distance is of the order of the thickness of the DNA/Dap2+ film. However, conversion of the supercoiled configuration to the circular form does appreciably change the number of targets available to produce the linear form or the multiple damages.31 We therefore find the formation of DSBs to be linear with fluence (bottom curve, Figure 1) and no saturation in the formation of isolated base damage, as revealed by enzymatic digestion (Figure 5).
DNA DSBs are generally considered to play a dominant role in the induction of cell death by ionizing radiation.44,60,61 Both experimental and theoretical investigations have demonstrated that, below 15 eV, electrons may induce strand breaks in DNA via the formation of TNIs,6,28,62,63 which decay into the DEA channel or by autoionization; this latter channel can leave a subunit in a repulsive state, thus also causing fragmentation. At 10 eV, electrons break DNA strands essentially via core-excited resonances, i.e., the formation of a transient anion consisting of the incoming electron trapped by the positive electron affinity of an electronically excited state of a basic unit of DNA.6,62,64
In our previous work, we evaluated the contribution of different lesions induced by γ-radiation to the loss of pGEM-3Zf (–) plasmid DNA functionality.42 We found that frank DSBs, which can induce cell lethality if not repaired, make a relatively minor contribution to the loss of DNA functionality induced by γ-radiation.42 Similarly, in the present study the frank DSBs induced by LEE irradiation are too infrequent to account for the loss of plasmid functionality. At a fluence of 22 × 1013 electrons/cm2, 49% of the plasmids were nonfunctional. Importantly, only 3% of the plasmids contained a DSB at this fluence (Figure 1). Thus, DSB formation cannot explain the loss of functionality of the plasmid observed here with LEEs.
Clustered lesions (i.e., local multiply damaged sites), represent a signature of the effects of ionizing radiation on DNA21 as a result of the occurrence of multiple damage events within one or two turns of the double-stranded helix along the particle track. However, assuming a Poisson distribution for the non-DSB cluster damaged plasmid (data are not shown), our analysis shows that at a fluence of 22 × 1013 electrons/cm2, 8% of the plasmids contained non-DSB cluster lesions. Since, at this fluence, 49% of the plasmids were nonfunctional, there must be additional lesions generated by LEEs, which are refractory to repair and therefore, lethal. The yield for total non-DSB cluster damage is about 2.6-fold higher than that of frank DSBs ((3.9 ± 0.6) × 10−16 cm2/electron vs (1.5 ± 0.2) × 10−16 cm2/electron). However, the yield for lethal damage βR is 31-fold higher than the yield of the non-DSB cluster damage ((1.2 ± 0.1) × 10−14 cm2/electron vs (3.9 ± 0.6) × 10−16 cm2/electron). In contrast, when we irradiated the DNA plasmid in aqueous solution with γ-radiation, the value for βR was only 2.6-fold greater than the yield of non-DSB cluster damage (βR = (60 ± 2) × 10−4 per Gy vs (22.9 ± 0.7) × 10−4 per Gy).42 This difference between the ratios of the yield of lethal damage to that of non-DSB cluster damage induced by LEEs and γ-radiation indicates that in these two different types of experiments, the energy of incident radiation is deposited within DNA very differently. This energy deposition depends on a number of factors, which in the case of LEE irradiation, are electron fluence, damage cross section, energy and thermalization range. For a dilute solution of DNA irradiated with γ-rays, the significant parameter is essentially the reaction rate constant of OH radicals to produce cluster damage. The magnitude of the parameters determines if the damage results from multiple hits (i.e., from two or more LEE, OH or eaq interactions) or a single particle-target process. When more than a single interaction is needed to cause the damage, its magnitude becomes nonlinear with dose or fluence.
In the previous experiments, the linearity of γ-induced clustered lesions with dose was not obvious below 100 Gy and absent above.42 Moreover, the loss of DNA functionality was observed for doses ranging from 300 to 800 Gy. Hence, this loss caused by clustered damages was observed under high concentrations of OH radicals and solvated electrons. Under such conditions, most multiple base damages and strand breaks should be caused by the reaction of two or more of these species with DNA, within a distance of about 10 base pairs along the axes of the plasmid.
The present result in Figure 5 clearly shows that the yields of DSB and non-DSB lesions are linear as a function of fluence. In other words, the yields of these damages, which cause the loss of functionality seen in Figure 2, are the result of a single electron interaction with DNA. Thus, the reactions and the experimental conditions are quite different in the LEE and γ-rays experiments. This finding is not too surprising, since many experiments performed, under single LEE collision conditions, with thin films of plasmid DNA and oligonucleotides, revealed that a single 10 eV electron interaction can cause a DSB.6,28 An electron transfer mechanism has been invoked to account for this phenomenon.65 Briefly, the incoming electron excites a dissociative electron state of a phosphate group leading to C–O bond cleavage on one strand. Afterward, the electron transfers to the opposite strand causing similar C–O bond cleavage via DEA. A similar mechanism has been shown to occur within the same strand by Park et al.66 When incident on single-stranded DNA a 10 eV electron first forms a core-excited TNI on a base; afterward, the electron is transferred to the phosphate group causing DEA at the C–O bond and leaving the base in a dissociative electronically excited state.67,68 Such electron-transfer mechanisms act over distances of the order of the wavelength of the captured electron.7,69 Thus, we expect multiple damages caused by a 10 eV electron to be extremely short-range and limited to distances of the order two base pairs.
It has been shown that repair enzymes, such as Fpg and Nth are unable to detect lesions in close proximity. Thus, we may have underestimated the frequency of non-DSB cluster damage created by LEEs. Indeed, the detection efficiency of cluster damage depends on the enzymatic conversion efficiency.70 Two factors can influence the detection of cluster damages by Fpg and Nth: the distance between two SSBs on opposite strands and the distance between base lesions on opposite strands of the DNA recognizable by these two enzymes.70 Complex lesions have been shown to be poorer substrates for these enzymes than single lesions.71 Where the lesions were only 1–3 bp apart on opposite strands, enzymatic digestion yielded only SSB; however, where they were 1–5 or 1–7 apart, they yielded DSBs.71 These findings were explained by postulating that a closely positioned break in the opposite strand inhibited the glycosylase activity of the enzyme, but not its lyase activity.71 Thus, the complex and compact lesions induce by LEEs should not be easily repaired and detected by comparison with γ-ray damage, not only in our experiment, but also by the enzyme repair machinery of the cells. As such LEE-induced clustered lesions could inactivate cell function or be converted to lethal damages such as DSBs by being recognized by the nucleotide excision repair (NER) system. DNA interstrand cross-links (ICLs) can also be responsible for the loss of plasmid DNA functionality, and have been reported to be converted to DSBs during repair.72–74 Some studies have found that a single ICL is sufficient to kill repair deficient bacteria.75–79 Recently, Price et al. demonstrated that ICL can be easily formed between adenine residues and abasic sites in duplex DNA in remarkably high yields (15–70%) under physiologically relevant conditions.80
SUMMARY AND CONCLUSION
The present work demonstrates that LEEs, which are produced in copious amounts by IR, are able to induce the loss of plasmid DNA functionality. DSBs are known to be toxic, but the yields induced by LEE impact ((3.9 ± 0.6) × 10−16 cm2/electron) is far too low to account for the loss of plasmid functionality. The yield for total non-DSB cluster damage was about 2.6 times higher than that of frank DSBs ((3.9 ± 0.6) × 10−16 vs (1.5 ± 0.2) × 10−16 cm2/electron), but is still too low to explain the toxicity of LEE irradiation. We suggest that LEEs induce compact cluster lesions that cannot be efficiently revealed by purified repair enzymes such as Fpg and Nth, thus leading to underestimation of non-DSB cluster lesions. The compact cluster damage, which is known to be poorly detected by repair enzymes in vitro, may also be poorly repaired in cells.
In conclusion, LEEs efficiently induce the loss of plasmid DNA functionality by generating lesions, which may be refractory to repair. The exact nature of these lesions remains unknown, but they seem to be compact clustered lesions. Further work is therefore needed to determine more precisely the effects of LEEs on DNA. It would be interesting to extend these measurements in order to determine the sensitivity of bacteria to electron damage at other energies and the role of TNIs in the formation of cluster lesions. Such knowledge is essential for understanding the details of energy deposition in cells by IR and should lead to the development of more efficient protocols for cancer treatment with radiotherapy.81,82 Such details may be particularly valuable for heavy ion therapy, where very high LET particles generate along their path a high density of LEEs,83 which, according to the present results, have a propensity to create less reparable cluster damages.
Acknowledgments
Financial support for this work was provided by the Canadian Institute of Health Research (Grant No. MOP 81356).
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Cobut V, Frongillo Y, Patau JP, Goulet T, Fraser M, Jay-Gerin J. Monte Carlo Simulation of Fast Electron and Proton Tracks in Liquid Water I. Physical and Physicochemical Aspects. Radiat Phys Chem. 1998;51:229–243. [Google Scholar]
- 2.Boudaïffa B, Cloutier P, Hunting D, Huels MA, Sanche L. Resonant Formation of DNA Strand Breaks by Low-Energy (3 to 20 eV) Electrons. Science. 2000;287:1658–1660. doi: 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- 3.Pimblott SM, LaVerne JA. Production of Low-Energy Electrons by Ionizing Radiation. Radiat Phys Chem. 2007;76:1244–1247. [Google Scholar]
- 4.Scifoni E, Surdutovich E, Solov’Yov AV. Spectra of Secondary Electrons Generated in Water by Energetic Ions. Phys Rev E. 2010;81:021903-1–7. doi: 10.1103/PhysRevE.81.021903. [DOI] [PubMed] [Google Scholar]
- 5.Kaplan IG, Miterev AM. The Delocalization of the Energy of the Ionizing Radiation in a Molecular Medium and its Radiation-Chemical Features. Radiat Phys Chem. 1985;26:53–56. [Google Scholar]
- 6.Sanche L. Low-Energy Electron Interaction with DNA: Bond Dissociation and Formation of Transient Anions, Radicals and Radical Anions. In: Greenberg M, editor. Wiley Series on Reactive Intermediates in Chemistry and Biology entitled Radicals in Nucleic Acids. John Wiley & Sons; Hoboken, NJ: 2009. pp. 239–293. [Google Scholar]
- 7.Barrios R, Skurski P, Simons J. Mechanism for Damage to DNA by Low-Energy Electrons. J Phys Chem B. 2002;106:7991–7994. [Google Scholar]
- 8.Baccarelli I, et al. Electron-Induced Damage of DNA and its Components: Experiments andtheoretical models. Phys Rep. 2011;508(1–2):1–44. [Google Scholar]
- 9.Ptasinska S, Sanche L. Dissociative Electron Attachment to Hydrated Single DNA Strands. Phys Rev E. 2007;75:031915-1–5. doi: 10.1103/PhysRevE.75.031915. [DOI] [PubMed] [Google Scholar]
- 10.Pan X, Cloutier P, Hunting D, Sanche L. Dissociative Electron Attachment to DNA. Phys Rev Lett. 2003;90:208102/1–208102/4. doi: 10.1103/PhysRevLett.90.208102. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Aleksandrov A, Orlando TM. Probing low-energy electron induced DNA damage using single photo-ionization mass spectrometry. Int J Mass Spectrom. 2008;277:314–320. [Google Scholar]
- 12.Ward JF. DNA Damage Produced by Ionizing Radiation in Mammalian Cells: Identities, Mechanisms of Formation, and Reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- 13.Little JB. Radiation Carcinogenesis. Carcinogenesis. 2000;21:397–404. doi: 10.1093/carcin/21.3.397. [DOI] [PubMed] [Google Scholar]
- 14.Little JB. Radiation-Induced Genomic Instability and Bystander Effects: Implications for Radiation Protection. Radio-protection. 2002;37:261–282. [Google Scholar]
- 15.Rydberg B, Lobrich M, Cooper PK. DNA Double-Strand Breaks Induced by High-Energy Neon and Iron Ions in Human Fibroblasts. I. Pulsed-Field Gel Electrophoresis Method. Radiat Res. 1994;139:133–141. [PubMed] [Google Scholar]
- 16.Trevor KT, Hersh EM, Brailey J, Balloul J, Acres B. Transduction of Human Dendritic Cells with a Recombinant Modified Vaccinia Ankara Virus Encoding MUC1 and IL-2. Cancer Immunol Immunother. 2001;50:397–407. doi: 10.1007/s002620100214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan HS. DNA-Strand Scission and Loss of Viability After X Irradiation of Normal and Sensitized Bacterial Cells. Proc Natl Acad Sci US A. 1966;55:1442–1446. doi: 10.1073/pnas.55.6.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonura T, Smith KC, Kaplan HS. Enzymatic Induction of DNA Double Strand Breaks in Irradiated Escherichia Coli K 12. Proc Natl Acad Sci US A. 1975;72:4265–4269. doi: 10.1073/pnas.72.11.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bresler SE, Noskin LA, Kuzovleva NA, Noskina IG. The Nature of the Damage to Escherichia coli DNA Induced by γ-Irradiation. Int J Radiat Biol. 1979;36:289–300. doi: 10.1080/09553007914551061. [DOI] [PubMed] [Google Scholar]
- 20.Schulte-Frohlinde D. Biological Consequences of Strand Breaks in Plasmid and Viral DNA. Br J Cancer. 1987;55:129–134. [PMC free article] [PubMed] [Google Scholar]
- 21.Gulston M, de Lara C, Jenner T, Davis E, O’Neill P. Processing of Clustered DNA Damage Generates Additional Double-Strand Breaks in Mammalian Cells Post-Irradiation. Nucleic Acids Res. 2004;32:1602–1609. doi: 10.1093/nar/gkh306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered Damages and Total Lesions Induced in DNA by Ionizing Radiation: Oxidized Bases and Strand Breaks. Biochemistry (N Y) 2000;39:8026–8031. doi: 10.1021/bi9927989. [DOI] [PubMed] [Google Scholar]
- 23.Sutherland BM, Bennett PV, Saparbaev M, Sutherland JC, Laval J, Preston RJ. Clustered DNA Damages as Dosemeters for Ionising Radiation Exposure and Biological Responses. Radiat Prot Dosim. 2001;97:33–38. doi: 10.1093/oxfordjournals.rpd.a006635. [DOI] [PubMed] [Google Scholar]
- 24.Sutherland BM, Bennett PV, Schenk H, Sidorkina O, Laval J, Trunk J, Monteleone D, Sutherland J. Clustered DNA Damages Induced by High and Low LET Radiation, Including Heavy Ions. Phys Medica. 2001;17:202–204. [PubMed] [Google Scholar]
- 25.Sutherland BM, Bennett PV, Sutherland JC, Laval J. Clustered DNA Damages Induced by X Rays in Human Cells. Radiat Res. 2002;157:611–616. doi: 10.1667/0033-7587(2002)157[0611:cddibx]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 26.Georgakilas AG, Bennett PV, Wilson DM, III, Sutherland BM. Processing of Bistranded Abasic DNA Clusters in γ-Irradiated Human Hematopoietic Cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mee LK, Adelstein SJ. Predominance of Core Histones in Formation of DNA-Protein Crosslinks in Gamma-Irradiated Chromatin. Proc Natl Acad Sci US A. 1981;78:2194–2198. doi: 10.1073/pnas.78.4.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huels MA, Boudaïffa B, Cloutier P, Hunting D, Sanche L. Single, Double, and Multiple Double Strand Breaks Induced in DNA by 3–100 eV Electrons. J Am Chem Soc. 2003;125:4467–4477. doi: 10.1021/ja029527x. [DOI] [PubMed] [Google Scholar]
- 29.Mirsaleh-Kohan N, Bass AD, Sanche L. Effect of morphology of thin DNA films on the electron stimulated desorption of anions. J Chem Phys. 2011;134(1):015102. doi: 10.1063/1.3520562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alizadeh E, Sanche L. Measurements of G-values for DNA damage induced by low energy electrons. J Phys Chem B. 2011;115(49):14852–14858. doi: 10.1021/jp207922n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rezaee M, Cloutier P, Bass AD, Michaud M, Hunting DJ, Sanche L. Absolute Cross Section for Low-Energy-Electron Damage to Condensed Macromolecules: A Case Study of DNA. Phys Rev E. 2012;86(301):031913-1–031913-10. doi: 10.1103/PhysRevE.86.031913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass AD, Sanche L. Interaction of Low Energy Electrons with Atomic and Molecular Solids. Marcel Dekker; New York: 2004. [Google Scholar]
- 33.Bass AD, Sanche L. Charge Trapping by H2O Condensed Onto Thin Films of Kr and Xe. J Chem Phys. 1991;95:2910–2918. [Google Scholar]
- 34.Boulanouar O, Khatyr A, Herlem G, Palmino F, Sanche L, Fromm M. Soft Adsorption of Densely Packed Layers of DNA-Plasmid 3 1,3-Diaminopropane Complexes Onto Highly Oriented Pyrolitic Graphite Designed to Erode in Water. J Phys Chem C. 2011;115:2191–2198. [Google Scholar]
- 35.Pegg AE. Recent Advances in the Biochemistry of Polyamines in Eukaryocytes. Biochem J. 1986;234:249–262. doi: 10.1042/bj2340249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spotheim-Maurizot M, Davídková M. Radiation Damage to DNA-Protein Complexes. J Phys Conf Ser. 2011;261:41–48. doi: 10.1016/j.mrfmmm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Newton GL, Aguilera JA, Ward JF, Fahey RC. Effect of Polyamine-Induced Compaction and Aggregation of DNA on the Formation of Radiation-Induced Strand Breaks: Quantitative Models for Cellular Radiation Damage. Radiat Res. 1997;148:272–284. [PubMed] [Google Scholar]
- 38.Iacomino G, Picariello G, Stillitano I, D’Agostino L. Nuclear Aggregates of Polyamines in a Radiation-Induced DNA Damage Model. Int J Biochem Cell Biol. 2014;47:11–19. doi: 10.1016/j.biocel.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Boudaïffa B, Cloutier P, Hunting D, Huels MA, Sanche L. Cross Sections for Low-Energy (10–50 eV) Electron Damage to DNA. Radiat Res. 2002;157:227–234. doi: 10.1667/0033-7587(2002)157[0227:csflee]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Panajotovic R, Martin F, Cloutier P, Hunting D, Sanche L. Effective Cross Sections for Production of Single-Strand Breaks in Plasmid DNA by 0.1 to 4.7 eV Electrons. Radiat Res. 2006;165:452–459. doi: 10.1667/rr3521.1. [DOI] [PubMed] [Google Scholar]
- 41.Orlando TM, Oh D, Chen Y, Aleksandrov AB. Low-Energy Electron Diffraction and Induced Damage in Hydrated DNA. J Chem Phys. 2008;128(19):195102-1–6. doi: 10.1063/1.2907722. [DOI] [PubMed] [Google Scholar]
- 42.Kouass Sahbani S, Girouard S, Cloutier P, Sanche L, Hunting JD. The relative contributions of DNA strand breaks, base damage and clustered lesions to the loss of DNA functionality induced by ionizing radiation. Radiat Res. 2013;181(1):99–110. doi: 10.1667/RR13450.1. [DOI] [PubMed] [Google Scholar]
- 43.Hanahan D. Studies on Transformation of Escherichia Coli with Plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 44.McMahon SJ, Currell FJ. A Robust Curve-Fitting Procedure for the Analysis of Plasmid DNA Strand Break Data from Gel Electrophoresis. Radiat Res. 2011;175:797–805. doi: 10.1667/RR2514.1. [DOI] [PubMed] [Google Scholar]
- 45.Zimmermann U, Neil G, editors. Electrotransformation of Bacteria, Electromanipulation of Cells. CRC Press; Boca Raton, FL: 1996. p. 110. [Google Scholar]
- 46.Ohse M, Kawade K, Kusaoke H. Effects of DNA Topology on Transformation Efficiency of Bacillus Subtilis ISW1214 by Electroporation. Biosci Biotechnol Biochem. 1997;61:1019–1021. doi: 10.1271/bbb.61.1019. [DOI] [PubMed] [Google Scholar]
- 47.Miller EM, Nickoloff JA. Escherichia coli Electro-transformation, Methods in Molecular Biology. Vol. 47. Humana Press; Totowa, NJ: 1995. pp. 105–113. [DOI] [PubMed] [Google Scholar]
- 48.Rezaee M, Alizadeh E, Hunting D, Sanche L. DNA-Platinum Thin Films for use in Chemoradiation Therapy Studies. Bioinorg Chem Appl. 2012;2012:923914-1–9. doi: 10.1155/2012/923914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cai Z, Cloutier P, Sanche L, Hunting D. DNA Interduplex Crosslinks Induced by AlKa X Rays Under Vacuum. Radiat Res. 2005;164:173–179. doi: 10.1667/rr3408. [DOI] [PubMed] [Google Scholar]
- 50.Dizdaroglu M, Laval J, Boiteux S. Substrate Specificity of the Escherichia coli Endonuclease III: Excision of Thymine- and Cytosine-Derived Lesions in DNA Produced by Radiation-Generated Free Radicals. Biochemistry. 1993;32:12105–12111. doi: 10.1021/bi00096a022. [DOI] [PubMed] [Google Scholar]
- 51.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New Substrates for Old Enzymes. 5-Hydroxy-2′-Deoxycytidine and 5-Hydroxy- 2′-Deoxyuridine are Substrates for Escherichia coli Endonuclease III and Formamidopyrimidine DNA N-Glycosylase, while 5-Hydroxy-2′-Deoxyuridine is a Substrate for Uracil DNA N-Glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 52.Purkayastha S, Milligan JR, Bernhard WA. On the Chemical Yield of Base Lesions, Strand Breaks, and Clustered Damage Generated in Plasmid DNA by the Direct Effect of X Rays. Radiat Res. 2007;168:357–366. doi: 10.1667/RR0964.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milligan JR, Aguilera JA, Wu CCL, Ng JY, Ward JF. The Difference that Linear Energy Transfer Makes to Precursors of DNA Strand Breaks. Radiat Res. 1996;145:442–448. [PubMed] [Google Scholar]
- 54.Milligan JR, Aguilera JA, Nguyen TD, Paglinawan RA, Ward JF. DNA Strand-Break Yields After Post-Irradiation Incubation with Base Excision Repair Endonucleases Implicate Hydroxyl Radical Pairs in Double-Strand Break Formation. Int J Radiat Biol. 2000;76:1475–1483. doi: 10.1080/09553000050176234. [DOI] [PubMed] [Google Scholar]
- 55.Hempel K, Mildenberger E. Determination of G-Values for Single and Double Strand Break Induction in Plasmid DNA using Agarose Gel Electrophoresis and a Curve-Fitting Procedure. Int J Radiat Biol. 1987;52:125–138. doi: 10.1080/09553008714551551. [DOI] [PubMed] [Google Scholar]
- 56.Gulston M, Fulford J, Jenner T, De Lara C, O’Neill P. Clustered DNA Damage Induced by γ Radiation in Human Fibroblasts (HF19), Hamster (V79–4) Cells and Plasmid DNA is Revealed as Fpg and Nth Sensitive Sites. Nucleic Acids Res. 2002;30:3464–3472. doi: 10.1093/nar/gkf467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokoya A, Cunniffe SMT, O’Neill P. Effect of Hydration on the Induction of Strand Breaks and Base Lesions in Plasmid DNA Films by γ-Radiation. J Am Chem Soc. 2002;124:8859–8866. doi: 10.1021/ja025744m. [DOI] [PubMed] [Google Scholar]
- 58.Milligan AJ, Metz JA, Leeper DB. The Effect of Lucanthone on Sublethal Radiation Damage, in Vivo. Int J Radiat Oncol Biol Phys. 1984;10:2309–2313. doi: 10.1016/0360-3016(84)90238-4. [DOI] [PubMed] [Google Scholar]
- 59.Durand RE, Brown SM. Effects of Lucanthone on Chinese Hamster V-79 Cells. I. Interaction with Radiation in Monolayers and Spheroids. Int J Radiat Oncol Biol Phys. 1980;6:1525–1530. doi: 10.1016/0360-3016(80)90010-3. [DOI] [PubMed] [Google Scholar]
- 60.Jeggo PA, Löbrich M. Contribution of DNA Repair and Cell Cycle Checkpoint Arrest to the Maintenance of Genomic Stability. DNA Repair. 2006;5:1192–1198. doi: 10.1016/j.dnarep.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 61.Psonka K, Gudowska-Nowak E, Brons S, Elsässer T, Heiss M, Taucher-Scholz G. Ionizing Radiation-Induced Fragmentation of Plasmid DNA—Atomic Force Microscopy and Biophysical Modeling. Adv Space Res. 2007;39:1043–1049. [Google Scholar]
- 62.Luo X, Zheng Y, Sanche L. DNA Strand Breaks and Crosslinks Induced by Transient Anions in the Range 2–20 eV. J Chem Phys. 2014;140:155101-1–11. doi: 10.1063/1.4870519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caron LG, Sanche L. Low-Energy Electron Diffraction and Resonances in DNA and Other Helical Macromolecules. Phys Rev Lett. 2003;91:1132011–1132014. doi: 10.1103/PhysRevLett.91.113201. [DOI] [PubMed] [Google Scholar]
- 64.Arumainayagam CR, Lee H, Nelson RB, Haines DR, Gunawardane RP. Low-Energy Electron-Induced Reactions in Condensed Matter. Surf Sci Rep. 2010;65:1–44. [Google Scholar]
- 65.Sanche L. Radiation Damage in Biomolecular Systems; Biological and Medical Physics, Biomedical Engineering. Springer; New York: 2012. Nanoscale Dynamics of Radiosensitivity: Role of Low Energy Electrons; pp. 3–43. [Google Scholar]
- 66.Park Y, Polska K, Rak J, Wagner JR, Sanche L. Fundamental Mechanisms of DNA Radiosensitization: Damage Induced by Low-Energy Electrons in Brominated Oligonucleotide Trimers. J Phys Chem B. 2012;116:9676–9682. doi: 10.1021/jp304964r. [DOI] [PubMed] [Google Scholar]
- 67.Zheng Y, Cloutier P, Hunting DJ, Sanche L, Wagner JR. Chemical Basis of DNA Sugar-Phosphate Cleavage by Low-Energy Electrons. J Am Chem Soc. 2005;127:16592–16598. doi: 10.1021/ja054129q. [DOI] [PubMed] [Google Scholar]
- 68.Zheng Y, Sanche L. Low Energy Electrons in Nanoscale Radiation Physics: Relationship to Radiosensitization and Chemo-radiation Therapy. Rev Nanosci Nanotechnol. 2013;2:1–28. [Google Scholar]
- 69.Caron L, Sanche L, Tonzani S, Greene CH. Low-Energy Electron Scattering from DNA Including Structural Water and Base-Pair Irregularities. Phys Rev A. 2009;80:012705-1–6. [Google Scholar]
- 70.Lomax ME, Gulston MK, O’Neill P. Chemical Aspects of Clustered DNA Damage Induction by Ionising Radiation. Radiat Prot Dosim. 2002;99:63–68. doi: 10.1093/oxfordjournals.rpd.a006840. [DOI] [PubMed] [Google Scholar]
- 71.Chaudhry MA, Weinfeld M. Induction of Double-Strand Breaks by S1 Nuclease, Mung Bean Nuclease and Nuclease P1 in DNA Containing Abasic Sites and Nicks. Nucleic Acids Res. 1995;23:3805–3809. doi: 10.1093/nar/23.19.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Halliwell B, Gutteridge JMC, Cross CE. Free Radicals, Antioxidants, and Human Disease: Where are we Now? J Lab Clin Med. 1992;119:598–620. [PubMed] [Google Scholar]
- 73.Noll DM, McGregor Mason T, Miller PS. Formation and Repair of Interstrand Cross-Links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sczepanski JT, Jacobs AC, Van Houten B, Greenberg MM. Double-Strand Break Formation during Nucleotide Excision Repair of a DNA Interstrand Cross-Link. Biochemistry. 2009;48:7565–7567. doi: 10.1021/bi901006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lawley PD, Brookes P. Molecular Mechanism of the Cytotoxic Action of Difunctional Alkylating Agents and of Resistance to this Action. Nature. 1965;206:480–483. doi: 10.1038/206480a0. [DOI] [PubMed] [Google Scholar]
- 76.Magana-Schwencke N, Pegas Henriques JA, Chanet R, Moustacchi E. The Fate of 8-Methoxypsoralen Photoinduced Crosslinks in Nuclear and Mitochondrial Yeast DNA: Comparison of Wild-Type and Repair-Deficient Strains. Proc Natl Acad Sci US A. 1982;79:1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duval-Valentin G, Takasugi M, Hélèene C, Sage E. Triple Helix-Directed Psoralen Crosslinks are Recognized by Uvr(A)BC Excinuclease. J Mol Biol. 1998;278:815–825. doi: 10.1006/jmbi.1998.1728. [DOI] [PubMed] [Google Scholar]
- 78.Truglio JJ, Croteau DL, van Houten B, Kisker C. Prokaryotic Nucleotide Excision Repair: The UvrABC System. Chem Rev. 2006;106:233–252. doi: 10.1021/cr040471u. [DOI] [PubMed] [Google Scholar]
- 79.Gregoli S, Olast M, Bertinchamps A. Radiolytic Pathways in -Irradiated DNA: Influence of Chemical and Conformational Factors. Radiat Res. 1982;89:238–254. [PubMed] [Google Scholar]
- 80.Price NE, Johnson KM, Wang J, Fekry MI, Wang Y, Gates KS. Interstrand DNA-DNA Cross-Link Formation between Adenine Residues and Abasic Sites in Duplex DNA. J Am Chem Soc. 2014;136:3483–3490. doi: 10.1021/ja410969x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tippayamontri T, Kotb R, Paquette B, Sanche L. Efficacy of Cisplatin and Lipoplatin in Combined Treatment with Radiation of a Colorectal Tumor in Nude Mouse. Anticancer Res. 2013;33:3005–3014. [PubMed] [Google Scholar]
- 82.Meesat R, et al. Cancer Radiotherapy Based on Femtosecond IR Laser-Beam Filamentation Yielding Ultra-High Dose Rates and Zero Entrance Dose. Proc Natl Acad Sci USA. 2012;109(38):E2508–E2513. doi: 10.1073/pnas.1116286109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hanin L, Zaider M. On the Probability of Cure for Heavy-Ion Radiotherapy. Phys Med Biol. 2014;59:3829–3842. doi: 10.1088/0031-9155/59/14/3829. [DOI] [PubMed] [Google Scholar]