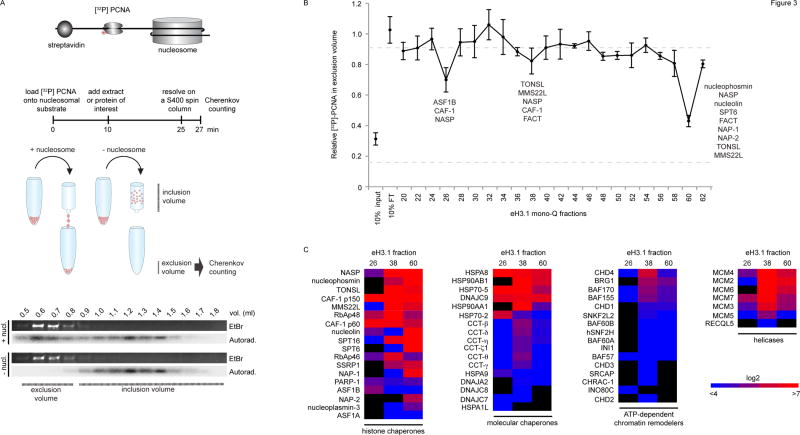
Figure 3.
Nucleosome disassembly. (A) A radiolabeled PCNA probe is loaded onto a linear DNA template, flanked by a single nucleosome at one extremity and a biotin-streptavidin block at the other. DNA flows into the exclusion volume when applied onto a gel filtration spin column, along with the radiolabeled PCNA trapped by the nucleosome. The radioactive probe is free to slide on DNA but falls off and remains in the inclusion volume if histones are removed. The assay quantifies the amount of radioactive PCNA in the exclusion volume by Cherenkov counting. (B) Testing nucleosome disassembly activity within affinity-purified eH3.1 from nuclear fractions (Figure 2). Data represents mean [32P]-PCNA retention +/− SD. (C) Mass spectrometry analysis (peptide counts) of the three fractions exhibiting histone eviction.