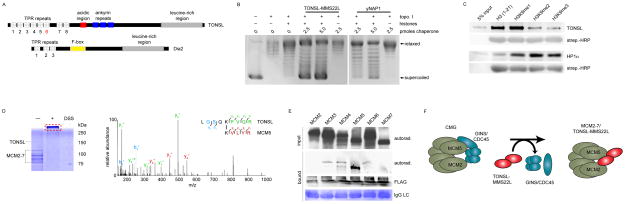
Figure 5.
TONSL is a histone chaperone that binds H3K9me1. (A) Primary structures of human TONSL and the similar yeast protein, Dia2. (B) Supercoiling assay demonstrating the histone chaperone ability of TONSL. (C) Pull-down assay utilizing immobilized histone peptides testing binding preference by recombinant TONSL and HP1. (D) MS/MS HCD spectrum of the (M + H)+4 ion of cross-linked peptides between TONSL (TRP repeat) and the C-terminus of MCM5. N-terminal fragment ions (b) are indicated in blue and C-terminal fragment ions (y) are indicated in green and red. The mass accuracy for precursor ion is better than 1 ppm and mass accuracy of all the fragment ions is better than 10 ppm. (E) Immobilized TONSL binds to in vitro translated MCM5. (F) Model for TONSL-MMS22L at the replication fork: Upon recruitment to stalled replication forks TONSL-MMS22L may maintain the CMG helicase inactive by binding to MCM5.