Abstract
Previous studies have demonstrated that bone marrow mesenchymal stromal cells (BMMSCs) from patients with myelodysplastic syndromes (MDS) display defective proliferative potential and impaired osteogenic differentiation ability. However, the underlying mechanisms are unclear. In the present study, the impaired osteogenic differentiation potential of BMMSCs was found in cases with RARS (83.3%), RCMD (75.0%), RAEB I (44.4%), RAEB II (40%). We also observed that MDS-BMMSCs with impaired osteogenic differentiation potential exhibited accelerate senescence and decreased hematopoietic supporting function. Further, we found that an abnormal activation of Notch-Hes signaling pathway in MDS-BMMSCs. By overexpression of Notch intracellular domain (NICD) in BMMSCs from healthy donors, we confirmed that Notch signaling pathway negatively regulated BMMSCs osteogenesis through inhibition of Runx2 transcriptional activity. Importantly, treatment with DAPT, a γ-secretase inhibitor of Notch signaling reversed the osteogenic differentiation in MDS-BMMSCs. Collectively, we provide evidence that activation of Notch-Hes signaling pathway is involved in the impaired osteogenic differentiation of MDS-BMMSCs and support the concept of a primary BMMSCs defect that might have a contributory effect in MDS pathogenesis.
Keywords: Bone marrow mesenchymal stromal cells, osteogenic differentiation, notch-hes pathway, myelodysplastic syndromes
Introduction
Myelodysplastic syndromes (MDS) are a heterogeneous group of clonal hematologic malignancies characterized by clonal dysplasia of hematopoietic stem/progenitor cells, leading to ineffective hematopoiesis and high risk of developingleukemia [1]. The abnormal of bone marrow microenvironment-the so called niche- is a very important factor in the pathogenesis of MDS. Bone marrow mesenchymal stromal cells (BMMSCs) are the main components in the niche with a critical role in the regulation of normal hematopoietic stem cells (HSCs) and their diverse progeny. Several groups have reported that BMMSCs from MDS patients were characterized as reduced proliferative capacity, increased senescence, impaired osteogenic differentiation ability, and diminished ability to support HSCs in long-term culture [2,3]. In our previous study, we also demonstrated that MDS-BMMSCs (mainly in lower-risk groups) had impaired osteogenic differentiation ability and reduced Runx2-positive osteoblast in bone marrow biopsy [4]. However, the molecular mechanisms associated with these abnormalities in MDS-BMMSCs are not understood well.
Notch signaling is a highly conserved signaling pathway associated with cell-fate determination, self-renewal potential, and apoptosis [5,6]. Notch receptors (Notch1-4) and their ligands (Jagged1-2 and Delta-like1,3,4) are families of transmembrane proteins with large extracellular domains. Both are expressed in BMMSCs. Notch signaling pathway has also been reported to be involved in the regulation of osteogenic differentiation of BMMSCs [7,8]. When Notch interacts with membrane-bound ligands on the surface of neighboring cells, γ-secretase-dependent cleavage of the Notch intracellular domain (NICD) will occur. The NICD then translocates into the nucleus, where it interacts with the CSL family of transcriptional regulators and initiates transcription of target genes such as Hes and Hey which inhibit Runx2, a transcription factor involved in osteogenic differentiation [9].
Notch signaling activation have been found in a variety of hematological malignancies and nonhematologic cancers, such as T cell acute lymphoblastic lymphomas/leukemias (T-ALL) and chronic lymphocytic leukemia (CLL), and is correlated with the prognostic and progression of these diseases [10]. However, the precise mechanism by which the Notch signaling pathway contributes to the pathogenesis of MDS remains elusive. Therefore, the aim of this study is to evaluate the osteogenic differentiation ability of BMMSCs from patients with MDS and to explore the role of Notch signaling pathway in the osteogenic differentiation of BMMSCs and the disturbed stromal function observed in patients with MDS.
Materials and methods
Patients
67 patients with MDS (median age 61 years, aged from 46 to 71) and 22 (median age 59 years, age from 43 to 69) normal controls were investigated in this study. All patients were untreated when they were recruited into this study. MDS was diagnosed in accordance with the minimum diagnostic criteria established by the Conference on MDS (Vienna, 2006) [11]. Assignment to different groups was decided according to the 2008 World Health Organization (WHO) classification [12] and the International Prognostic Scoring System (IPSS) [13]. All patients were classified for the study as “lower-risk (LR)” (IPSS-low/int-1), and as “higher-risk(HR)” (IPSS-int-2/high). Detailed information about these MDS patients is presented in Table 1. All subjects provided informed consent. This study was approved by the Ethics Committee of the Sixth Hospital affiliated with Shanghai Jiao Tong University, and all patient relevant research strictly abided by the Declaration of Helsinki.
Table 1.
The clinical characteristic of all MDS patients
| Characteristic | ||
|---|---|---|
| Median age (range) | 61 (46-71) | |
| Sex | Male | 39 |
| Female | 28 | |
| WHO classification | RA | 3 |
| RARS | 6 | |
| RCMD | 32 | |
| RAEB I | 15 | |
| RAEB II | 11 | |
| IPSS | Lower risk (≤ 1) | 45 |
| Higher risk (> 1) | 22 |
IPSS, International Prognostic Scoring System; RA, refractory anaemia; RARS, refractory anaemia with ringedsideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RAEB, RA with excess blasts.
Isolation and culture of BMMSCs
The BM mononuclear cells (BMMNCs) were isolated by Ficoll-Paque Plus (GE Healthcare, Uppsala, Sweden). BMMNCs were seeded at an initial concentration of 1×106 cells/ml and cultured in Human Mesenchymal Stem Cell Growth Medium (Cyagen Biosciences Inc, Guangzhou, China) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2 in fully humidified atmosphere. The supernatant containing non-adherent cells was removed and replaced with fresh supplemented medium every 3-4 days. When the monolayer was established (80-90% confluence) , cells were detached with 0.25% trypsin-EDTA (Gibico), At the third passage (P3), BMMSCs were evaluated by flow cytometry for the absence of CD34, CD45 antigens and the presence of CD73, CD90, CD105 and CD166.
Cell growth assay
BMMSCs were seeded at 2000 cells/well in 96-well plate for 1-7 days. At different time points the cell number was measured using Cell-Counting Kit-8 (CCK8) proliferation assay kit (Beyotime, China). BMMSCs were mixed with 10 μl of CCK-8 solution/well and incubated for further 2 hours at 37°C. The amount of formazan dye generated by cellular dehydrogenase activity was measured for absorbance at 450 nm with a microplate reader. The optical density (OD) values of each well represented the survival/proliferation of BMMSCs.
SA-β-Gal assay
BMMSCs cultured on plates were washed with PBS and fixed in 4% paraformaldehyde for 15 min at room temperature. After rinsing with PBS, cells were incubated with a freshly prepared SA-β-Gal staining solution (Beyotime, China) for 16 h at 37°C. Under light microscopy, the number of blue cells (SA-β-Gal positive cells) out of at least 500 cells in 10 randomly chosen fields was used to calculate the percentage of senescent cells.
Osteogenic differentiation assay
BMMSCs were seeded at 3×104 cells/well in 6-well plate pre-coated with gelatin solution in Human Mesenchymal Stem Cell Osteogenic Differentiation Medium (Cyagen Biosciences Inc, Guangzhou, China), then the medium was replaced every 3-4 days. After 3 weeks differentiation, cells can be fixed and stained with Alizarin red, visualized using light microscopy. After photography, the bound staining was eluted with 10% (wt/vol) cetylpyridinium chloride (sigma), and alizarin red-S in samples was quantified by measuring absorbance at 572 nm. ALP activity was assessed using alkaline phosphatase activity kit (Jiancheng Biotechnology Institute, Nanjing, China), following the manufacturer’s instructions.
Hematopoietic assay
CD34+ cells were isolated by magnetic-activated cell sorting (MACS) (Miltenyi Biotec, Bergisch Gladbach, Germany) from BMMNCs. CD34+ cells purity was evaluated with FACS (BD Biosciences, NJ, USA) and was > 90%. 1.2×106 BMMSCs were cultivated on 96-well plates until at least 80% confluence was reached and then irradiated with 30 Gray. Afterwards, 1×104 CD34+ cells were plated on these BMMSCs feeder layers and cultivated in long term bone marrow culture (LTBMC) medium consisting of MyeloCult H5100 (Stem Cell Technologies, Vancouver, BC, Canada) supplemented with 10-6 mol/L hydrocortisone (Sigma) for 5 weeks with weekly changes of culture medium. After 5 weeks, the medium was replaced by clonogenic methylcellulose medium consisting of 50 ng/mL of SCF, 10 ng/mL of IL-3 (Sigma), 20 ng/mL of GM-CSF (Sigma), and 4 U/mL of erythropoietin (Sigma). After 2 weeks, hematopoietic colonies greater than 50 cells were counted.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was extracted from BMMSCs using the RNeasy Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions. cDNA was prepared using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Burlington, Canada) following the manufacturer’s protocol. To perform real-time PCR, each 20 μl RT-PCR mix contained 10 μl of RealMasterMix (Takara, Dalian, China), 0.8 μl of each prime, 2 μL cDNA, and distilled water. PCR was performed on an ABI 7500 real-time PCR machine (Applied Biosystems). The primer sequences of Notch-1, Notch-2, Deltalike-1, Jagged-1, hairy and Enhancer of split homolog-1 (Hes1), hairy and Enhancer of split homolog-2 (Hes2), hairy and Enhancer of split homolog-5 (Hes5), hes-related family bHLH transcription factor with YRPW motif 1 (Hey1), hes-related family bHLH transcription factor with YRPW motif 2 (Hey2), hes-related family bHLH transcription factor with YRPW motif L (HeyL), runt related transcription factor 2 (Runx2), Osterix, bone sialoprotein (BSP), alkaline phosphatase (ALP), Type1 collagen (COL-1), osteopontin (OPN) and osteocalcin (OCN) are listed in Table 2. GAPDH served as reference control, and differences in mRNA expression levels were calculated as fold changes by the 2-ΔΔCt method.
Table 2.
The sequence of primers used for real time PCR
| Prime | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| GAPDH | CCCACTCCTCCACCTTTGA | CCACCCTGTTGCTGTAGCC |
| Notch1 | CTTGTGTCAACGGCGGC | TTGGGACCGCTGAAGCC |
| Notch2 | GGCCACCTGAAGGGAAGCACATA | CACAGAGGCTGGGAAAGGATGATA |
| Hes1 | AGGCTGGAGAGGCGGCTAAG | TGGAAGGTGACACTGCGTTGG |
| Hes5 | ACCAGCCCAACTCCAAGCT | GGCTTTGCTGTGCTTCAGGTA |
| Hey1 | GGATCACCTGAAAATGCTGCATAC | CCGAAATCCCAAACTCCGATAG |
| Hey2 | GAACAATTACTCGGGGCAAA | TCAAAAGCAGTTGGCACAAG |
| HeyL | AGCCAGGAAGAAACGCAGAGG | GCTGTTGAGGTGGGAGAGAAGG |
| Deltalike1 | TCCTGATGACCTCGCAACAGA | ACACACGAAGCGGTAGGAGT |
| Jagged1 | TCGGGTCAGTTCGAGTTGGA | AGGCACACTTTGAAGTATGTGTC |
| Runx2 | AGTGGACGAGGCAAGAGTTTC | CCTTCTGGGTTCCCGAGGT |
| ALP | CCATTCCCACGTCTTCACATT | AAGGGCTTCTTGTCTGTGTCACT |
| COL1 | CACCAATCACCTGCGTACAGAA | CAGATCACGTCATCGCACAAC |
| BSP | GACAGTTCAGAAGAGGAGGAG | AGCCCAGTGTTGTAGCAGA |
| OCN | AGGGCAGCGAGGTAGTGAA | TCCTGAAAGCCGATGTGGT |
| OPN | TTTACAACAAATACCCAGATGC | ATGGCTTTCGTTGGACTTACT |
Western blot analysis
Whole cell lysates were obtained from BMMSCs and equal quantities of protein were separated by 12% SDS-PAGE and blotted onto a PVDF membrane. The PVDF membranes were blocked with Tris-buffered saline (TBS) containing 5% skimmed milk powder for 1 h, incubated with rabbit hes1 (Abcam Inc.), rabbit Runx2 (Abcam Inc.) and mouse GAPDH (Abcam Inc) as loading control. Membranes were incubated with either anti-mouse or anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences). Specific bands were visualized by using ECL Western Blotting Detection Reagents (Amersham Biosciences).
Treatment with the Notch1 inhibitor DAPT
MDS-BMMSCs were cultured in osteogenic induction medium containing the Notch1 inhibitor N-[N-(3,5-Difluorophenacetyl)-l-alanyl]-S-phenylglycinet-butyl Ester (DAPT, 5 nM; Sigma, Saint Louis, MO, USA), or vehicle (dimethyl sulfoxide, DMSO; Sigma, Saint Louis, MO, USA) for 48 hours.
Generation of lentivirus vector constructs and transduction
The adenovirus expression vector that encodes human NICD was generated by using the Adeno-X Expression System (BD Biosciences Clontech). The recombinant virus was packaged and amplified in HEK293 cells and purified by CsCl density gradient centrifugation. To package adenovirus, the adenoviral vectors were linearized with the restriction enzyme PacI and transfected into HEK293 cells using Lipofectamine2000. After several rounds of propagation, recombinant adenovirus was purified by an AdEasy virus purification kit (Stratagene). The transfection was performed according to the manufacturer’s protocol.
Statistical analysis
The data were presented as mean ± SD. All statistical analyses were performed using the SPSS 17.0 System. Comparison of mRNA levels between healthy controls and different MDS subtypes was using a Student’s t test, oneway analysis of variance (ANOVA) was used to assess multiple pairwise comparisons. p < 0.05 was considered statistically significant.
Results
Decreased osteogenic differentiation in BMMSCs of MDS patients
The first step of our study comprised the evaluation of the osteoblastic differentiation potential of BMMSCs from MDS patients and healthy controls. After 21 days of osteogenic induction, Alizarin red staining was used to visualize osteogenic differentiation. Relative calcium production by BMMSCs from MDS subtypes (RARS 5/6, RCMD 14/16, RAEB I 4/9, RAEB II 2/5) patients was reduced after 21 days of differentiation when compared to healthy controls (Figure 1A, 1B). mRNA expression of Runx2 was determined using RT-PCR without osteogenic induction in BMMSCs, the results showed that Runx2 was significantly reduced in BMMSCs from some MDS subtypes (RARS, RCMD) (p < 0.05) (Figure 1C).
Figure 1.
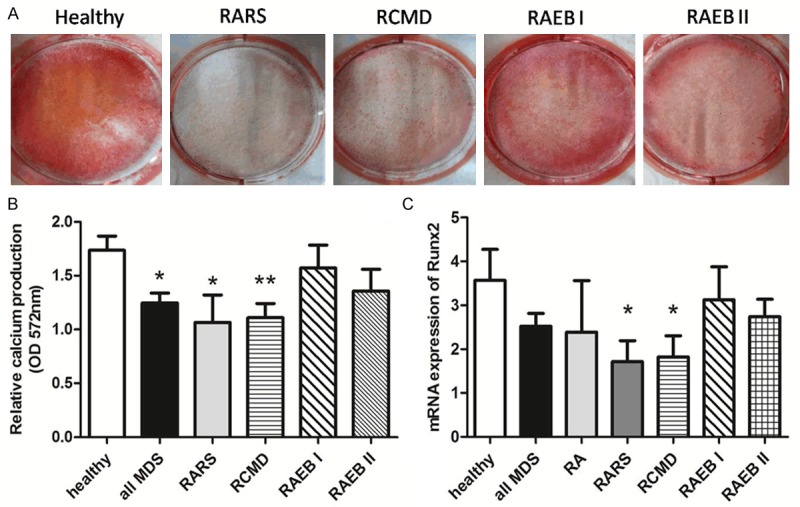
Impaired osteogenic differentiation potential of MDS-BMMSCs. A. After 21 days of osteogenic induction, Alizarin red staining was used to visualize osteogenic differentiation. Representative micrographs of BMMSCs derived from healthy controls (HC) and different MDS subtypes are shown. B. Relative calcium production (OD 572 nm) by BMMSCs from some MDS subtypes (all MDS n = 36, RARS n = 6, RCMD n = 16), patients was reduced after 21 days of differentiation when compared to healthy controls (HC n = 11), and some MDS subtypes BMMSCs (RAEB I n = 9, RAEB II n = 5) showed no difference when compared with healthy controls BMMSCs. C. mRNA expression of Runx2 was determined using RT-PCR without osteogenic induction in BMMSCs of healthy controls (HC n = 22) and MDS (all MDS n = 67, RA n = 3, RARS n = 6, RCMD n = 32, RAEB I n = 15, RAEB II n = 11). The results showed that mRNA expression of Runx2 was significantly reduced in BMMSCs from some MDS subtypes (RARS, RCMD). The results were expressed as means ± SD. Compared with HC-BMMSCs, significance was set as *p < 0.05, **p < 0.01.
MDS-BMMSCs with impaired osteogenic differentiation were prone to senescence, accompanied by decreased proliferation and stem cell-supporting capacity
To further analyze the biological characteristics of MDS-BMMSCs which have poor osteogenic differentiation potential. MDS-BMMSCs were divided into two groups: well-osteo MDS-BMMSCs (relative calcium production OD 572 nm value ≥ 1.5) and impaired-osteo MDS-BMMSCs (relative calcium production OD 572 nm value < 1.5). The degree of osteogenic potential was estimated by relative calcium production after 21 days of osteogenic induction. SA-β-Gal was used to examine BMMSCs senescence, the cell count revealed that the number of SA-β-gal-positive cells from impaired-osteo MDS-BMMSCs were significantly higher than normal BMMSCs and well-osteo MDS-BMMSCs (p < 0.05 ) (Figure 2A, 2B). The proliferation of BMMSCs was measured with CCK-8 assay, BMMSCs from impaired-osteo MDS patients grew more slowly than those from the other groups (p < 0.05 ) (Figure 2C). To assess the ability of hematopoietic support by BMMSCs from MDS patients, colony-forming units (CFU) such as Colony forming unit-granulocyte, macrophage (CFU-GM), Burst forming unit-erythroid (BFU-E) and Colony forming unit-granulocyte, erythrocyte, macrophage, megakaryocyte (CFU-GEMM) were counted. The results showed that the number of CFU-GM, BFU-E and CFU-GEMM on MDS-derived BMMSCs was significantly less than that of normal-BMMSCs (p < 0.05) (Figure 2D).
Figure 2.
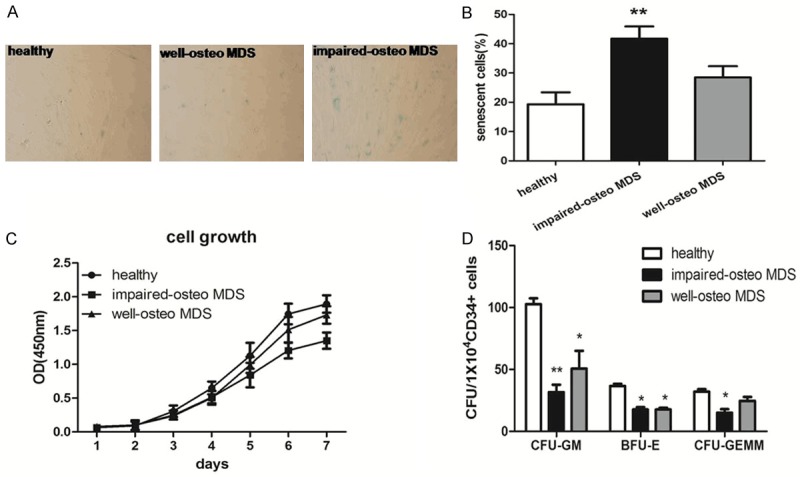
SA-β-Gal activity, proliferation and stem cell-supporting capacity of MDS-BMMSCs. MDS-BMMSCs were divided into two groups: well-osteo MDS-BMMSCs (relative calcium production OD 572 nm value ≥ 1.5, n = 11) and impaired-osteo MDS-BMMSCs (relative calcium production OD 572 nm value < 1.5, n = 25). A, B. Cellular senescence was assessed by SA-β-Gal staining in BMMSCs of normal controls and MDS patients. The number of blue cells (SA-β-Gal positive cells) out of at least 500 cells in 10 randomly chosen fields was used to calculate the percentage of senescent cells. The mean percentage of SA-β-Gal positive cells is higher in impaired-osteo MDS-BMMSCs compared to normal BMMSCs. C. Growth curve of BMMSCs was tested by CCK-8 assay. The absorbance was shown as the proliferation rate. BMMSCs from impaired-osteo MDS patients grew more slowly than those from the control group and well-osteo MDS. D. BMMSCs were cultured in LTBMC medium with CD34+ cells for 5 weeks followed by methylcellulose progenitor culture for additional 2 weeks. The number of CFU-GM, BFU-E and CFU-GEMM on MDS-derived BMMSCs was significantly less than that of normal-BMMSCs. The results were expressed as means ± SD. Compared with HC-BMMSCs, significance was set as *p < 0.05, **p < 0.01.
Up-regulation of the Notch-Hes signaling pathway in BMMSCs of MDS patients
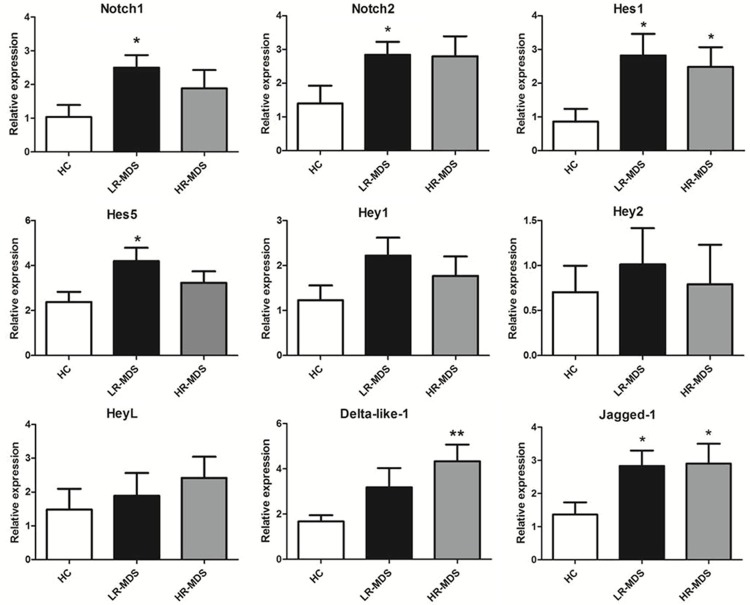
To determine whether the impaired osteogenic differentiation ability of BMMSCs in MDS patients was associated with abnormal Notch signaling pathway, we investigated the expression of genes implicated in the Notch signaling (including Notch ligands Jagged-1 and Delta-like-1, Notch receptors Notch1 and Notch2, Notch signaling downstream genes hes1, hes5, hey1, hey2 and heyL) by RT-PCR. The results showed that mRNA expression of Jagged-1, Notch1, Notch2, hes1 and hes5 was significantly increased in BMMSCs from LR-MDS compared to healthy controls (p < 0.05 ). The mRNA expression of hes1, Jagged-1, and Delta like-1 was significantly upregulated in BMMSCs from HR-MDS (p < 0.05 ) (Figure 3).
Figure 3.
Relative mRNA expression of Notch signaling ligands, receptors and downstream genes in MDS-BMMSCs. RT-PCR was performed in detecting the mRNA expression levels of Notch signaling ligands (Jagged-1 and Delta like-1), receptors (Notch1, Notch2) and downstream genes (hes1, hes5, hey1, hey2 and heyL). MDS-BMMSCs were divided into two groups: lower-risk MDS (LR-MDS n = 45) and higher-risk-MDS (HR-MDS n = 22). The results showed that mRNA expression of Jagged-1, Notch1, Notch2, hes1 and hes5 was significantly increased in BMMSCs from LR-MDS compared to healthy controls (HC n = 22). The mRNA expression of hes1, Jagged-1 and Delta like-1 was significantly upregulated in BMMSCs from HR-MDS. The results were expressed as means ± SD. Compared with HC-BMMSCs, significance was set as *p < 0.05, **p < 0.01.
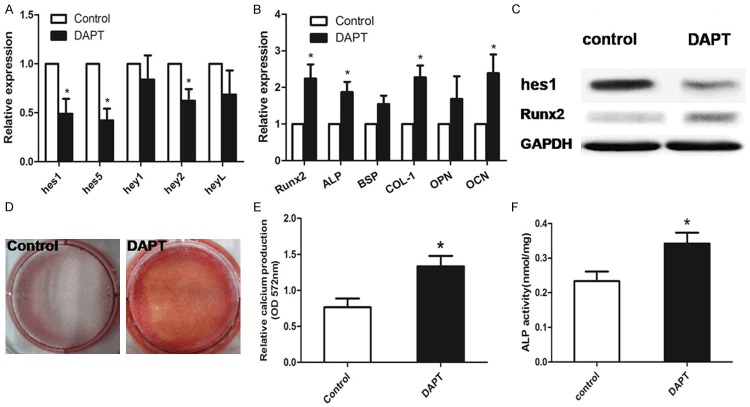
Treatment with DAPT reversed the osteogenic differentiation in MDS-BMMSCs with impaired osteogenic differentiation
To further assess whether Notch signaling was functionally involved in the osteogenic commitment of BMMSCs from MDS patients. We used DAPT, a γ-secretase inhibitor to inhibit Notch activation. Addition of 10 mM DAPT in growth medium for 48 h reduced the level of hes1, hes5 and hey2 expression for MDS-BMMSCs with impaired osteogenic differentiation (Figure 4A). Impaired-osteo MDS-BMMSCs showed increased mRNA expression of osteogenic differentiation related genes (Runx2, ALP, COL-1, OCN) after culturing in osteogenic induction medium supplemented with DAPT for 7 days (Figure 4B). In order to confirm quantitative PCR data, Western blot analysis was performed. After DAPT treatment, BMMSCs from impaired-osteo MDS patients showed a lower protein expression of hes1 and a higher protein expression of Runx2 (Figure 4C), supporting gene expression results. In line with the gene expression, impaired-osteo MDS-BMMSCs treated with DAPT demonstrated increased mineralization after 21 days of osteogenic induction and enhanced ALP activity after 3 days of osteogenic induction (p < 0.05) (Figure 4D, 4F).
Figure 4.
DAPT-treated MDS-BMMSCs with impaired osteogenic differentiation. A. Addition of 10 mM DAPT in growth medium for 48 h reduced the level of (hes1, hes5, hey2) expression for MDS-BMMSCs with impaired osteogenic differentiation. B. Impaired-osteo MDS-BMMSCs showed increased mRNA expression of osteogenic differentiation related genes (Runx2, ALP, COL-1, OCN) after culturing in osteogenic induction medium supplemented with DAPT for 7 days. C. Inhibition of hes1 production by DAPT significantly enhanced the protein levels of Runx2. D, E. Impaired-osteo MDS-BMMSCs treated with DAPT demonstrated increased mineralization assessed by alizarin red staining after 21 days of osteogenic induction. F. DAPT-treated impaired-osteo MDS-BMMSCs showed enhanced ALP activity after 3 days of osteogenic induction. The results were expressed as means ± SD, significance was set as *p < 0.05.
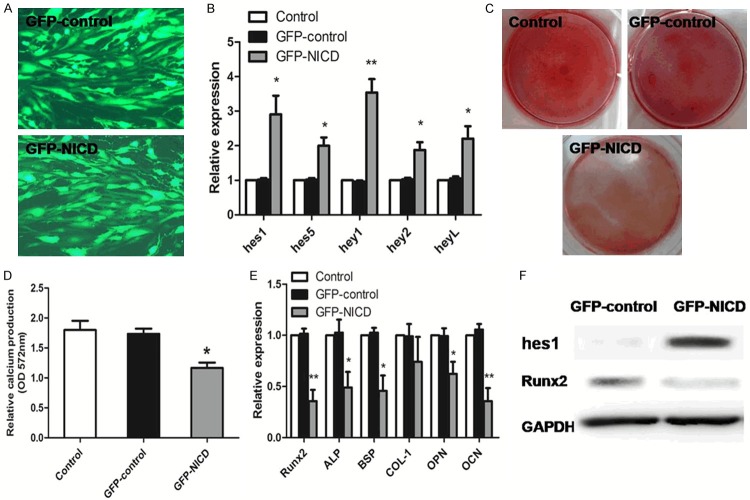
Overexpression of NICD in normal-BMMSCs impaired osteogenesis
In the following experiment, we used normal-BMMSCs transfected with adenovirus carrying Notch intracellular domain (NICD) (GFP-NICD) or GFP (GFP-control). hes1 (the key gene of Notch signaling downstream genes) expression was considerably increased in the GFP-NICD BMMSCs at both the mRNA (Figure 5B) and protein (Figure 5F) levels (p < 0.05). The mRNA expression of other Notch target genes (hes5, hey1, hey2 and heyL) were also upregulated in the GFP-NICD BMMSCs (p < 0.05) (Figure 5B). After 21 days of osteogenic induction, relative calcium production by NICD adenovirus transfection cells was reduced after 21 days of differentiation when compared to GFP control-transduced cells (Figure 5C, 5D). The osteogenic differentiation related genes (Runx2, BSP, ALP, OPN, OCN) was reduced in NICD adenovirus transfection cells compared to GFP control-transduced cells after 7 days of osteogenic induction differentiation (p < 0.05) (Figure 5E). Overexpression of NICD significantly downregulated the protein levels of Runx2 (Figure 5F).
Figure 5.
Overexpression of notch1 intracellular domain (NICD) of normal-BMMSCs. A. Transfection efficiency under the fluorescence microscope. B. The mRNA expression of Notch target genes (hes1, hes5, hey1, hey2 and heyL) demonstrates upregulated expression in NICD adenovirus transfection expressing cells compared to the GFP control-transduced cells. C. After 21 days of osteogenic induction, Alizarin red staining was used to visualize osteogenic differentiation. Representative micrographs of normal-BMMSCs, GFP control-transduced BMMSCs and NICD adenovirus transfection BMMSCs. D. Relative calcium production by NICD adenovirus transfection cells was reduced after 21 days of differentiation when compared to GFP control-transduced cells. E. The osteogenic differentiation related genes (Runx2, BSP, ALP, OPN, OCN) was reduced in NICD adenovirus transfection cells compared to GFP control-transduced cells after 7 days of osteogenic induction differentiation. F. Overexpression of NICD significantly downregulated the protein levels of Runx2. The results were expressed as means ± SD. Compared with GFP control-BMMSCs, significance was set as *p < 0.05, **p < 0.01.
Discussion
Recent research showed that deletion of Dicer1 (miRNA processing endonuclease) from osteoprogenitor cells led to an impaired osteogenic differentiation of BMMSCs and induced an MDS phenotype in an animal model [14], suggesting that alterations of the microenvironment alone might induce MDS. However, whether osteoprogenitor cell abnormalities are involved in the pathogenesis of MDS is still unknown. In our previous study, we found that there was downregulation of Dicer1 in MDS-BMMSCsandt impaired osteogenic differentiation potential existed largely in lower-risk-MDS patients [4,15], which were consistent with other studies [16,17]. To clarify the underlying pathogenetic mechanism, in the present study, we investigated the osteogenic differentiation potential and the activation of Notch signaling pathways of BMMSCs in MDS patients.
Our results confirmed that there was impaired osteogenic differentiation potential of BMMSCs in cases with RARS (83.3%), RCMD (75.0%), RAEB I (44.4%), RAEB II (40%), which was characterized as the downregulation of Runx2 mRNA expression and relative calcium production. To investigate the biological characteristics of MDS-BMMSCs with poor osteogenic differentiation potential, we divided MDS-BMMSCs into two groups: well-osteo BMMSCs and impaired-osteo BMMSCs. The data revealed that impaired osteogenic MDS-BMMSCs were prone to senescence, accompanied by decreased proliferation and damaged stem cell-supporting capacity. As we know, osteoblast represent a regulatory component of the BM niche, controlling the size of HSCs pool and HSCs quiescence [18,19]. The abnormal osteoblasts would directly affect the composition of BM niche. Thus, the impaired osteogenic differentiation potential of BMMSCs may be a contributing factor of the ineffective hematopoiesis in patients with MDS.
A growing body of evidence has shown that increased expression of Notch genes and their ligands are detected in many hematopoietic malignant cells [20,21]. However, little is known about the Notch signaling in MDS-BMMSCs. We have therefore studied the expression of several Notch signaling related genes in BMMSCs by RT-PCR. Among all tested genes, the expression of Notch ligands Delta-like-1, Jagged-1 were significantly up-regulated in MDS-BMMSCs compared to healthy controls. These results, which were consistent with results of Geyh et al. [16] and Varga et al. [22]. Since the over-expression of soluble Jagged1 on MDS-BMMSCs has been shown to result in a reduced rate of cobble-stone forming cells in MDS stromal coculture [22], raising a interesting possibility that the increased expression of Jagged-1 by MDS-BMMSCs could be involved in their defective hematopoiesis supporting capacity. Other studies also found that Delta-like-1 was strongly over-expressed in MDS BM trephines and BMMSCs [23,24]. Besides up-regulation of Notch ligands, we found the expression levels of several Notch receptors and downstream genes such as Notch1, Notch2, hes1, hes5 also altered in MDS-BMMSCs. The above evidences suggested that their deregulation contributes to the insufficient hematopoietic support of MDS-BMMSCs.
In bone marrow, Notch signaling plays a role in maintaining the pool of MSCs by suppressing osteogenic differentiation through inhibition of Runx2 transcriptional activity [10]. We may therefore hypothesize that the diminished osteogenic differentiation of MDS-BMMSCs might be associated with the activation of the Notch signaling. To confirm our hypothesis, DAPT, a γ-secretase inhibitor, was used to inhibit Notch activation [25]. The results showed that addition of DAPT reduced expression levels of hes1 and enhanced the expression of Runx2 in impaired osteo-MDS-BMMSCs. In addition, inhibition of Notch signaling enhanced the expression of osteogenic markers, and remarkably increased ALP activity and mineralization deposition compared with control cells. These results suggested that activation of notch signaling pathway was involved in the impaired osteogenic differentiation of MDS-BMMSCs and Notch inhibitor might be an effective approach to promote osteogenesis of MDS-BMMSCs. Our study also demonstrated the role of Notch signaling in the osteogenic differentiation of healthy controls-BMMSCs, we showed that activation of Notch signaling (through NICD) in these cells strongly impair their osteogenic differentiation potential characterized by a reduction in calcium deposits and the expression of osteoblastic maker genes. Consistent results were also found in multiple myeloma (MM) derived-BMMSCs, in which maintenance of Notch signaling activity was involved in a suppression of the osteogenic differentiation in MM-BMMSCs [8]. Interestingly, our study revealed that impaired osteogenic MDS-BMMSCs were prone to senescence. However, the activated Notch signaling could induce senescence of BMMSCs through the p53/p21 pathway [26], and senescent cells have harmful effects on the tissue microenvironment by secrete several pro-inflammatory cytokines [27,28]. These studies indicated that activated Notch signaling might be responsible for the abnormal microenvironment in MDS.
Notch signaling pathway is considered as an attractive target for treatment in cancer [29,30]. Notch modulating drugs are already in clinical trials [31]. Reducing Notch activity in cancer stem cells may promote their differentiation, thus reducing their ability to form the tumor mass [32]. Similarly, Notch inhibitor might be an effective approach to promote osteogenesis of BMMSCs and adjust BM niche. The specific stromal target therapy of MDS still needs further research.
In summary, our data demonstrate that activation of Notch signaling pathway is involved in the impaired osteogenic differentiation of MDS-BMMSCs and reflect an abnormal microenvironment that fails to support normal haematopoiesis. Our results support the theory that a primary stromal defect may have a contributory effect in MDS.
Acknowledgements
This study was supported in part by the National Natural Science Foundation of China (NNSFC81170463, NNSFC81400090).
Disclosure of conflict of interest
None.
References
- 1.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111:4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 2.Flores-Figueroa E, Montesinos JJ, Flores-Guzmán P, Gutiérrez-Espíndola G, Arana-Trejo RM, Castillo-Medina S, Pérez-Cabrera A, Hernández-Estévez E, Arriaga L, Mayani H. Functional analysis of myelodysplastic syndromes-derived mesenchymal stem cells. Leuk Res. 2008;32:1407–1416. doi: 10.1016/j.leukres.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Zhao ZG, Xu W, Yu HP, Fang BL, Wu SH, Li F, Li WM, Li QB, Chen ZC, Zou P. Functional characteristics of mesenchymal stem cells derived from bone marrow of patients with myelodysplastic syndromes. Cancer Lett. 2012;317:136–143. doi: 10.1016/j.canlet.2011.08.030. [DOI] [PubMed] [Google Scholar]
- 4.Fei C, Zhao Y, Gu S, Guo J, Zhang X, Li X, Chang C. Impaired osteogenic differentiation of mesenchymal stem cells derived from bone marrow of patients with lower-risk myelodysplastic syndromes. Tumour Biol. 2014;35:4307–4316. doi: 10.1007/s13277-013-1565-6. [DOI] [PubMed] [Google Scholar]
- 5.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 6.Duncan AW, Rattis FM, DiMascio LN, Congdon KL, Pazianos G, Zhao C, Yoon K, Cook JM, Willert K, Gaiano N, Reya T. Integration of Notch and Wnt signaling in hematopoietic stem cell maintenance. Nat Immunol. 2005;6:314–322. doi: 10.1038/ni1164. [DOI] [PubMed] [Google Scholar]
- 7.Hilton MJ, Tu X, Wu X, Bai S, Zhao H, Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R, Long F. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu S, Evans H, Buckle C, De Veirman K, Hu J, Xu D, Menu E, De Becker A, Vande Broek I, Leleu X, Camp BV, Croucher P, Vanderkerken K, Van Riet I. Impaired osteogenic differentiation of mesenchymal stem cells derived from multiple myeloma patients is associated with a blockade in the deactivation of the Notch signaling pathway. Leukemia. 2012;26:2546–2549. doi: 10.1038/leu.2012.126. [DOI] [PubMed] [Google Scholar]
- 9.Zamurovic N, Cappellen D, Rohner D, Susa M. Coordinated activation of Notch, Wnt and TGF-β signaling pathways in BMP-2 induced osteogenesis: Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem. 2004;279:37704–37715. doi: 10.1074/jbc.M403813200. [DOI] [PubMed] [Google Scholar]
- 10.Jundt F, Schwarzer R, Dorken B. Notch signaling in leukemias and lymphomas. Curr Mol Med. 2008;8:51–59. doi: 10.2174/156652408783565540. [DOI] [PubMed] [Google Scholar]
- 11.Valent P, Horny HP, Bennett JM, Fonatsch C, Germing U, Greenberg P, Haferlach T, Haase D, Kolb HJ, Krieger O, Loken M, van de Loosdrecht A, Ogata K, Orfao A, Pfeilstöcker M, Rüter B, Sperr WR, Stauder R, Wells DA. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leuk Res. 2007;31:727–736. doi: 10.1016/j.leukres.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100:2292–2302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, Ohyashiki K, Toyama K, Aul C, Mufti G, Bennett J. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 14.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, Aung Z, Matza M, Merkenschlager M, Lin C, Rommens JM, Scadden DT. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Wu D, Fei C, Guo J, Gu S, Zhu Y, Xu F, Zhang Z, Wu L, Li X, Chang C. Downregulation of Dicer1 promotes cellular senescence and decreases the differentiation and stem cellsupporting capacities of mesenchymal stromal cells in patients with myelodysplastic syndrome. Haematologica. 2015;100:194–204. doi: 10.3324/haematol.2014.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geyh S, Oz S, Cadeddu RP, Fröbel J, Brückner B, Kündgen A, Fenk R, Bruns I, Zilkens C, Hermsen D, Gattermann N, Kobbe G, Germing U, Lyko F, Haas R, Schroeder T. Insufficient stromal support in MDS results from molecular and functional deficits of mesenchymal stromal cells. Leukemia. 2013;27:1841–1851. doi: 10.1038/leu.2013.193. [DOI] [PubMed] [Google Scholar]
- 17.Ferrer RA, Wobus M, List C, Wehner R, Schönefeldt C, Brocard B, Mohr B, Rauner M, Schmitz M, Stiehler M, Ehninger G, Hofbauer LC, Bornhäuser M, Platzbecker U. Mesenchymal stromal cells from patients with myelodyplastic syndrome display distinct functional alterations that are modulated by lenalidomide. Haematologica. 2013;98:1677–85. doi: 10.3324/haematol.2013.083972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, Harris S, Wiedemann LM, Mishina Y, Li L. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 19.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 20.Weng AP, Ferrando AA, Lee W, Morris JP 4th, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 21.Kannan S, Sutphin RM, Hall MG, Golfman LS, Fang W, Nolo RM, Akers LJ, Hammitt RA, McMurray JS, Kornblau SM, Melnick AM, Figueroa ME, Zweidler-McKay PA. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med. 2013;210:321–337. doi: 10.1084/jem.20121527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga G, Kiss J, Várkonyi J, Vas V, Farkas P, Pálóczi K, Uher F. Inappropriate Notch activity and limited mesenchymal stem cell plasticity in the bone marrow of patients with myelodysplastic syndromes. Pathol Oncol Res. 2007;13:311–319. doi: 10.1007/BF02940310. [DOI] [PubMed] [Google Scholar]
- 23.Qi X, Chen Z, Liu D, Cen J, Gu M. Expression of Dlk1 gene in myelodysplastic syndrome determined by microarray, and its effects on leukemia cells. Int J Mol Med. 2008;22:61–68. [PubMed] [Google Scholar]
- 24.Länger F, Stickel J, Tessema M, Kreipe H, Lehmann U. Overexpression of delta-like (Dlk) in a subset of myelodysplastic syndrome bone marrow trephines. Leuk Res. 2004;28:1081–1083. doi: 10.1016/j.leukres.2004.01.020. [DOI] [PubMed] [Google Scholar]
- 25.Vujovic S, Henderson SR, Flanagan AM, Clements MO. Inhibition of γ-secretases alters both proliferation and differentiation of mesenchymal stem cells. Cell Prolif. 2007;40:185–195. doi: 10.1111/j.1365-2184.2007.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Huang L, Sun H. Role of Notch expression in premature senescence of murine bone marrow stromal cells. Prog Nat Sci. 2009;19:557–562. [Google Scholar]
- 27.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davalos AR, Coppe JP, Campisi J, Desprez PY. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 2010;29:273–283. doi: 10.1007/s10555-010-9220-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Zhou X, Redfield S, Lewin J, Miele L. Elevated Jagged-1 and Notch-1 expression in high grade and metastatic prostate cancers. Am J Transl Res. 2013;5:368–378. [PMC free article] [PubMed] [Google Scholar]
- 31.Krop I, Demuth T, Guthrie T, Wen PY, Mason WP, Chinnaiyan P, Butowski N, Groves MD, Kesari S, Freedman SJ, Blackman S, Watters J, Loboda A, Podtelezhnikov A, Lunceford J, Chen C, Giannotti M, Hing J, Beckman R, Lorusso P. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J. Clin. Oncol. 2012;30:2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 32.Evans AG, Calvi LM. Notch signaling in the malignant bone marrow microenvironment: implications for a niche-based model of oncogenesis. Ann N Y Acad Sci. 2015;1335:63–77. doi: 10.1111/nyas.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]