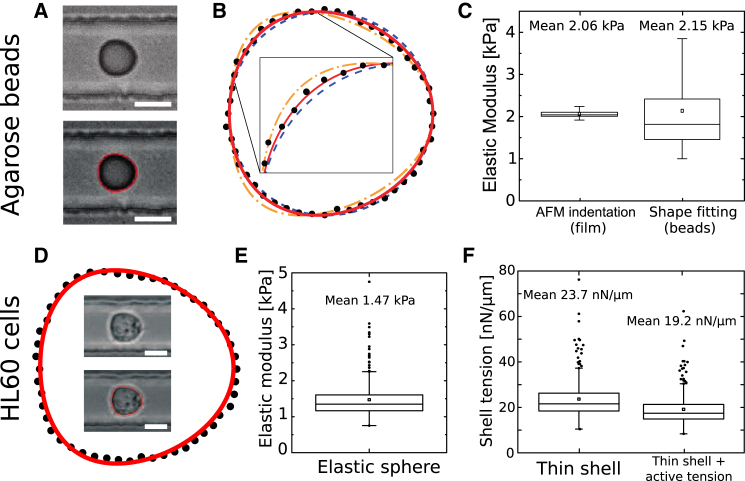
Figure 5.
Shape fitting of elastic agar beads and HL60 cells. (A) Deformed bead in the microfluidic channel (top left) overlaid with the contour found by the automatic contour detection (bottom left) (17). (B) Contour found by the tracking software (dotted line) overlaid by shapes from the theory for an elastic sphere for different elastic moduli, kPa (yellow dash-dotted line), kPa (red solid line, best fit, 95% confidence interval: 0.3 kPa) and kPa (blue dashed line). (C) Comparison of results from shape fitting of measured contours with AFM indentation on agar film of the same composition as the beads ( different spots indented). (D) Deformed cell in the microfluidic channel. In the outer contour, the cell boundary found by the tracking software is shown (black dots) overlaid by the shape of a solid elastic sphere of elastic modulus kPa according to our theory (red solid line). (E and F) Cell elastic moduli from shape fitting using the cell mechanical model of (E) a solid elastic sphere or (F) a thin elastic shell without (left) and with a relative surface tension of (right). All measurements were taken with μL/s, mPa·s. In (C), (E), and (F), box plots show the mean value (square), first to third quartiles (horizontal lines), and 1.5× the interquartile region (whiskers). In (E) and (F), outliers are shown in addition (black circles). Scale bars in (A) and (D) are 10 μm. To see this figure in color, go online.