Figure 7.
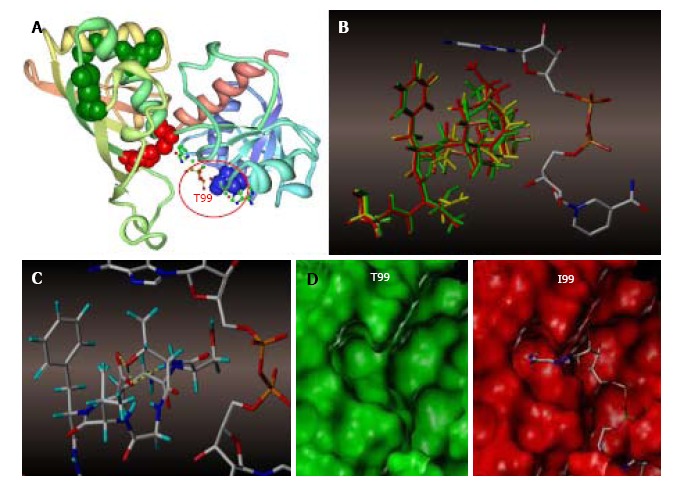
Molecular modeling of NAD+ binding site in human glyceraldehyde 3-phosphate dehydrogenase. A: NAD+ binding site within a GAPDH polypeptide (circled). Only one of four GAPDH subunits is shown. T99 is shown as a space-filled amino acid; B: The results of the trajectory calculations, with the average structure for each simulation shown overlaid for T99 (green), A99 (yellow), and I99 (red), along with the NAD+ molecule bound in the active cleft. Isoleucine side chain (red) of the T99I polypeptide acquires the closest position relative to the purine ring; C: In the native T99, an intramolecular H-bond between T99 and E97 (highlighted yellow) is likely to exist which holds the loop in a restricted conformation. This hydrogen bond could be a stabilizing factor permitting NAD+ to fully occupy the upper region of the cleft; D: Side-by-side comparison of T99 (green) and I99 (red) NAD+ binding site surfaces. In I99, the average structure places the terminus of the side chain well within the Van der Waals radii of the adenine ring of NAD+, thereby displacing or blocking the cofactor from its initial association with GAPDH. GAPDH: Glyceraldehyde 3-phosphate dehydrogenase.
