Figure 4.
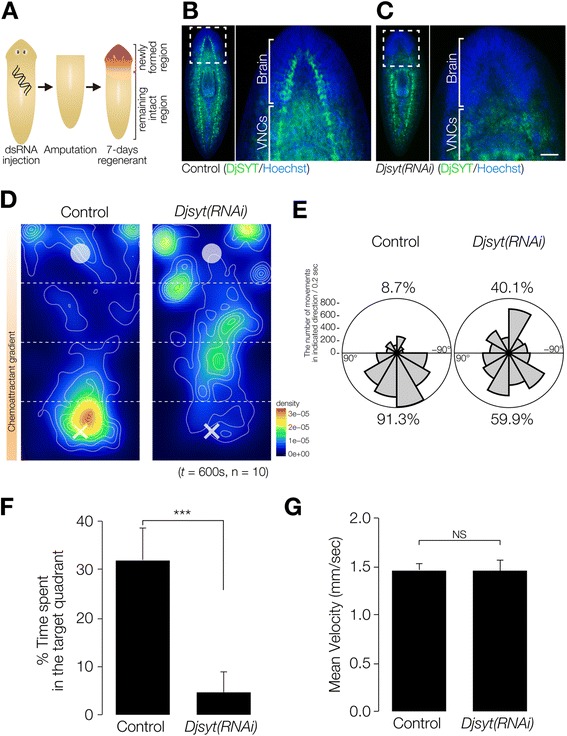
Chemotaxis of planarians that had lost of brain neural activity by Readyknock of synaptotagmin gene. (A) Schematic illustration of experimental design of Readyknock of synaptotagmin gene. After injection of double-stranded RNA of Djsyt, planarians were amputated, and then allowed to regenerate their heads for 7 days. Red-colored portion indicates newly regenerated head. (B, C) Control and Readyknock of Djsyt. Immunohistochemical detection of DjSYT protein, shown in green in control (B) and in Readyknock (C) animals 7 days after decapitation. Samples were stained with Hoechst 33342 (for nuclei, shown in blue) to visualize planarian tissues, including brain. The dashed boxes indicate the border between the newly formed head region magnified in the right panels. Readyknock using Djsyt(RNAi) treatment caused severe reduction of the level of DjSYT protein, although strong signals of the DjSYT protein were still detected in the pre-existing ventral nerve cords (VNCs) in the trunk region. Bar, 150 μm. (D) Heat map view with contour lines of chemotaxis of control and Djsyt(RNAi) planarians in chemoattractant concentration gradient field. Djsyt(RNAi) planarians showed random movement, whereas control planarians moved to and stayed in the region having the highest concentration of chemoattractant in the chemoattractant-gradient field. (E) Rose plots show orientation of movement of control and Djsyt(RNAi) planarians in Zones 2 and 3 in chemoattractant gradient-field. The movement of control planarians was biased toward being orientated toward the highest concentration of chemoattractant, whereas Djsyt(RNAi) animals in the chemoattractant gradient-field showed no particular orientation of movement. (F) Time spent in the target quadrant (Zone 4) during assay of control and Djsyt(RNAi) planarians in the chemoattractant gradient-field is shown as mean ± SEM. (G) Mean velocity mean of control and Djsyt(RNAi) planarians during assay. ***, p < 0.005; NS, not significant; t = 600 sec; n = 10.
