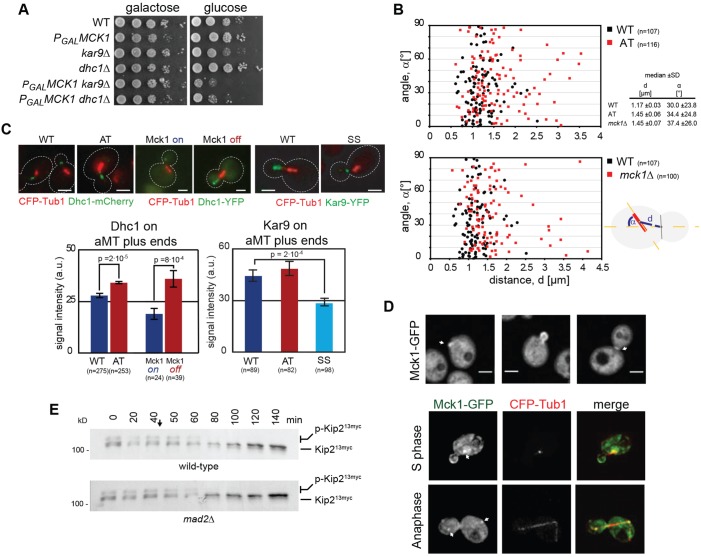
Fig. 6.
Kip2 phosphorylation controls the amount of Kar9 and dynein on aMT plus ends and spindle positioning. (A) Genetic interactions of MCK1 with genes required for spindle positioning. PGALHAMCK1 kar9Δ and PGALHAMCK1 dyn1Δ cells display slow growth upon deactivation of MCK1 expression on glucose-containing plates. (B) Spindles are mispositioned in mck1Δ cells and cells expressing Kip2-AT. Plotted is the angle of the spindle to the mother-bud axis (α, y-axis), against the distance from the bud neck (d, x-axis), the vertical line denotes the bud neck. Number of spindles counted in brackets. (C) Fluorescence intensity of Dyn1–YFP and Kar9–YFP on aMT plus ends in cells expressing the Kip2 variants indicated or in cells expressing wild-type Kip2 in HAMck1 overexpressing or depleted cells. Numbers of counted aMTs in brackets, arbitrary intensity units ±s.e.m. are shown. Scale bar: 3 µm. (D) Localisation of Mck1–GFP. A Mck1 sub-pool localises to the incipient bud, to the bud cortex until G2/M phase, and the bud neck. Mck1 also localises to the spindle poles (possibly SPBs or kinetochores) from S phase until anaphase. Scale bar: 2.5 µm. (E) Phosphorylation of Kip2 requires MTs. Time course showing Kip213myc phosphorylation in extracts of wild type- and spindle assembly checkpoint-defective mad2Δ cells, after MT depolymerisation. Nocodazole was added at 40 min (arrow). Equal number of cells were used to generate the extracts for each time point.