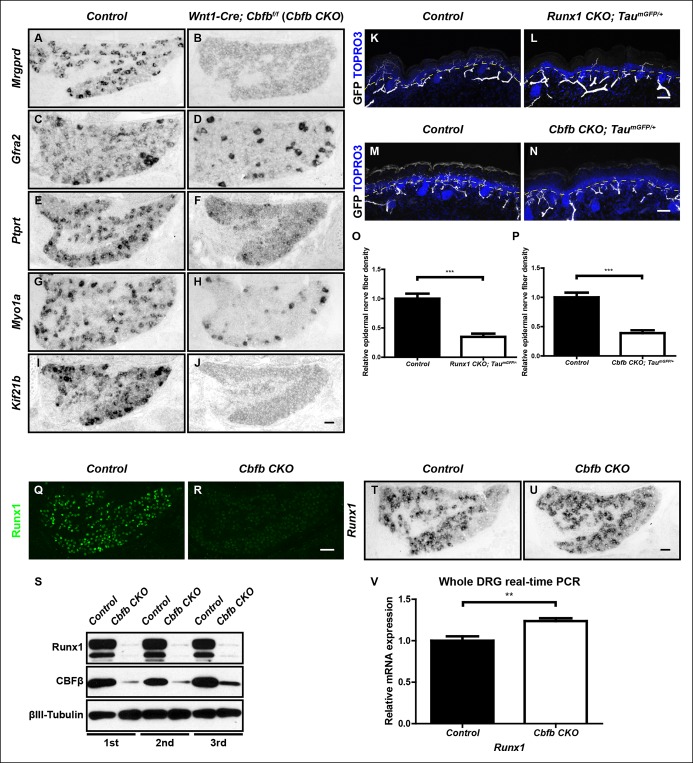
Figure 5. CBFβ is required for acquisition of molecular and morphological features of nonpeptidergic nociceptors.
(A–J) Expression of Mrgprd (Control, 26.9% ± 2.8%; Cbfb CKO, 0%), Gfra2 (Control, 38.8% ± 2.8%; Cbfb CKO, 11.7% ± 1.9%), Ptprt, (Control, 31.9% ± 3.2%; Cbfb CKO, 7.1% ± 2.8%), Myo1a (Control, 26.9% ± 3.2%; Cbfb CKO, 5.6% ± 0.6%) and Kif21b (Control, 20.2% ± 0.1%; Cbfb CKO, 2.4% ± 0.5%) in control and Cbfb CKO DRGs at P0 by in situ hybridization analysis. The gene expression deficits in Cbfb CKO animals phenocopy those observed in Runx1 CKO animals except for Kif21b expression. The discrepancy likely reflects Kif21b expression in proprioceptors where it presumably depends on Runx3 and CBFβ for expression. Shown are the means ± SEMs for the percentage of neurons expressing indicated genes based on counts from a total of at least 9 sections from three independent animals per genotype. DRG neurons were identified and counted based on combined NeuN immunostaining, which was not shown. See also Figure 5—figure supplement 1D, E. (K–N) GFP immunostaining of P0 hairy skin to visualize sensory innervation of the epidermis in control and Runx1 CKO animals (K and L) or control and Cbfb CKO animals (M and N) that also carry the TaumGFP allele. The TaumGFP allele was introduced to label all Cre-expressing neurons including all sensory neurons. Note that there is a dramatic reduction in fiber density specifically in the epidermis in both Runx1 CKO and Cbfb CKO animals relative to their littermate controls. The yellow dotted line denotes the epidermal-dermal junction which was drawn based on TOPRO3 counterstain (blue). (O and P) Quantification of sensory innervation of the epidermis in control and Runx1 CKO animals (O) or control and Cbfb CKO animals (P) reveals a remarkably similar reduction in the innervation density in both mutants at P0. The innervation density is defined as the fraction of area occupied by GFP+ fibers in the epidermis. An unpaired t test was performed on data from three independent animals per genotype. ***p ≤ 0.001. (Q and R) Runx1 immunostaining of control and Cbfb CKO DRGs at P0 shows almost complete loss of Runx1 proteins in the absence of CBFβ. Shown are representative images from at least three independent experiments. (S) Immunoblot analysis of expression of Runx1 and Cbfb in control and Cbfb CKO DRGs at P0 shows dramatic loss of Runx1 proteins as a result of CBFβ depletion. βIII-Tubulin serves as a loading control. Shown are results from three independent experiments. (T and U) In situ hybridization analysis of Runx1 expression in control and Cbfb CKO DRGs at P0 shows comparable levels of Runx1 transcripts in control and mutant animals. (V) Real-time PCR analysis of Runx1 expression in control and Cbfb CKO DRGs at P0 shows increased Runx1 mRNA expression in Cbfb CKO DRGs compared to control, which likely reflects an increased ratio of nociceptors to proprioceptors (data not shown). An unpaired t test was performed on data from four independent pairs of control and mutant animals, **p ≤ 0.01. Cbfbf/f mice were used as control animals for analysis of Cbfb CKO mutants. Scale bar, 50 μm.