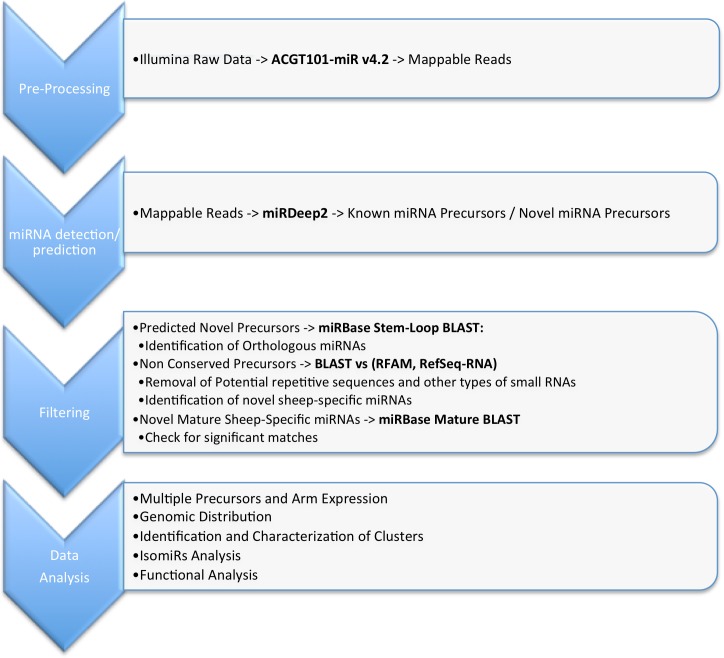
Fig 3. Computational pipeline of data analysis.
The figure illustrates the four steps of the computational pipeline employed to analyze the RNAseq data. Pre-Processing: raw data were processed by the ACGT101-miR v4.2 pipeline in order to obtain good quality mappable reads. miRNA detection/prediction: this step was carried out by applying miRDeep2 to the mappable reads. The tool returned the lists of known and novel miRNA precursors identified. Filtering: the output of miRDeep2 was further analyzed by BLAST against different databases in order to assess evolutionary conservation of the predicted miRNAs and remove sequences matching other kinds of small RNAs. Data Analysis: last step consisted of the application of a series of ad-hoc scripts for the extraction of descriptive statistics. The tool IPA was used to perform the functional enrichment analysis of the targets of the identified miRNAs, which were predicted by the software miRiam.