Fig. 5.
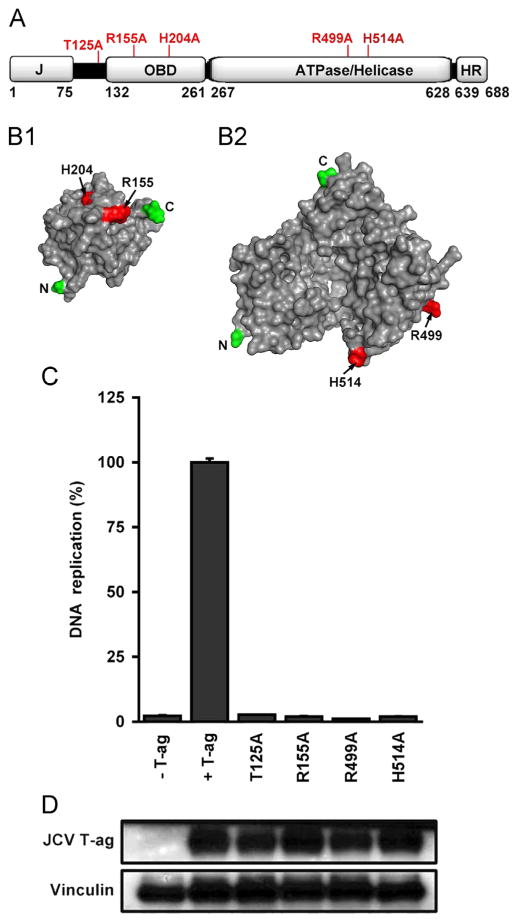
JCV DNA replication assays conducted with mutant forms of T-ag. A. A linear depiction of JCV large T-ag showing its major domains (i.e., J, OBD, ATPase/helicase and host range (HR)). The relative locations of the T-ag residues that were mutated to alanines (see section ‘Materials and methods’) are indicated on the top of the figure. B1. A surface representation of the JCV OBD (Meinke et al., 2014) showing the relative position of R155 and H204 (shown in red). B2. A surface representation of the JCV helicase domain, prepared using the program Phyre 2 (Kelley and Sternberg, 2009). This model shows the location of a residue at the base of the beta-hairpin (H514) and a residue that, based on studies of SV40 T-ag (Li et al., 2003), is likely involved in ATP binding (R499). C. JCV replication assays conducted in C33A cells for 72 h. with T-ag molecules containing mutations at residues that are predicted to be essential for DNA replication. The extreme left column presents the result of a replication reaction conducted in the absence of T-ag, while the amount of replication obtained with wt JCV T-ag is shown in the second column. The following four columns present the amount of replication obtained with the T125A, R155A, R499A and H514A mutants. D. Results from a Western blot demonstrating that levels of “mutant T-ag” are not different from those of wt JCV T-ag. As a loading control, levels of Vinculin were also determined.