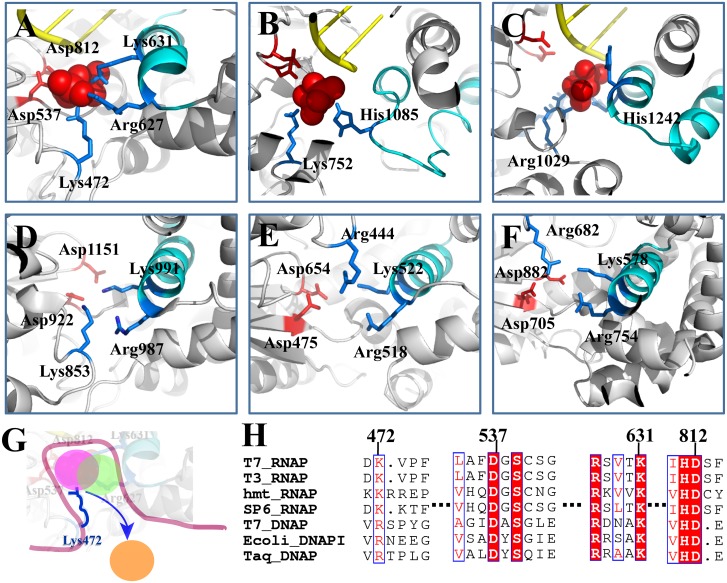
Fig 6. Comparing T7 RNAP with other RNAPs or DNAPs on the key PPi interactions around the active site. (A) T7 RNAP (PDB:1S77); (B) yeast Pol II [26]; (C) bacterial RNAP [25]; (D) human mitochondrial RNAP (PDB:3SPA); (E) T7 DNAP (PDB:1T7P); (F) E. coli DNAP I (PDB:1KLN).
(G) A schematic of the jump from the cavity model, depicting the key lysine/arginine module that assists the PPi release from the active site (the colored disks represent the three meta-stable states as identified for T7 RNAP); (H) The sequence alignment of the single-subunit polymerases based on similarities of special sequence motifs and molecular structures. The essential charged residues for the PPi release are labeled. The conservations of corresponding residues in the sequence alignment are highlighted.