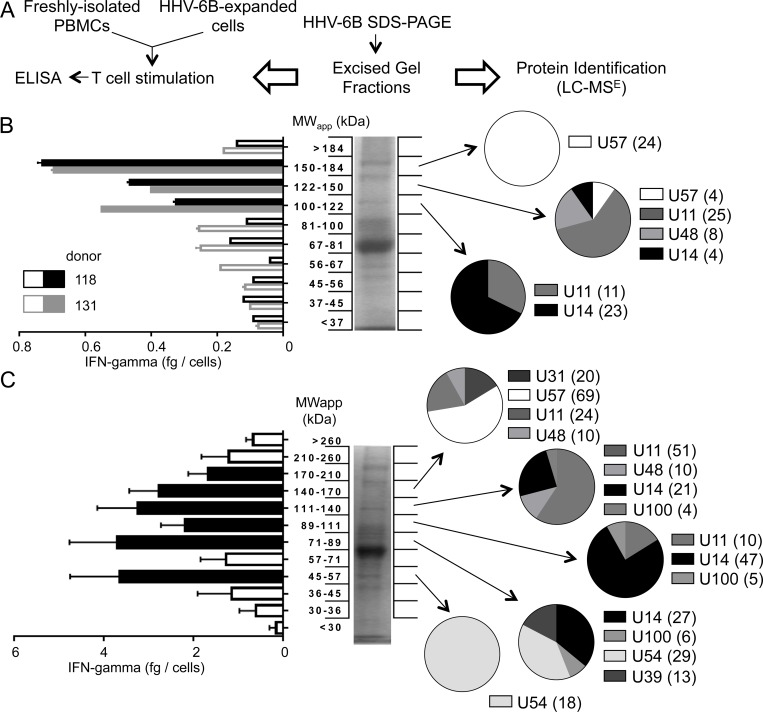
Fig 2. Identification of HHV-6B targets of the IFN-γ response.
A. Schematic of the experimental approach: fractionation of proteins by SDS-PAGE; measurement of IFN-γ secretion induced by gel fractions; identification of HHV-6B proteins by proteomics in selected fractions. B and C. IFN-γ production by PBMCs (B) or in vitro HHV-6B-expanded T cell cultures (C), in response to stimulation with gel fractions containing HHV-6B proteins. Middle panels show SDS-PAGE profiles of extracellular virus; left panels show IFN-γ production (fg/cell) by each fraction; right panels show protein identification by proteomics (pie charts show the numbers of tryptic peptides characterized by mass spectrometry used to identify each HHV-6B protein; numbers are shown in parentheses). In B, individual IFN-γ responses of two different donors are shown using black (#118) and grey (#131) bars, with filled bars indicating positive responses (values above the background mean + 3 times standard deviation). Error bars represent standard deviation of replicate measurements for each donor. Filled-bars indicate fractions selected for proteomics analysis. In C, average IFN-γ responses of four expanded T cell cultures (from two different donors); error bars represent the standard deviation. Filled bars indicate positive responses (values above the background mean + 3 times the standard deviation), selected for proteomics analysis.