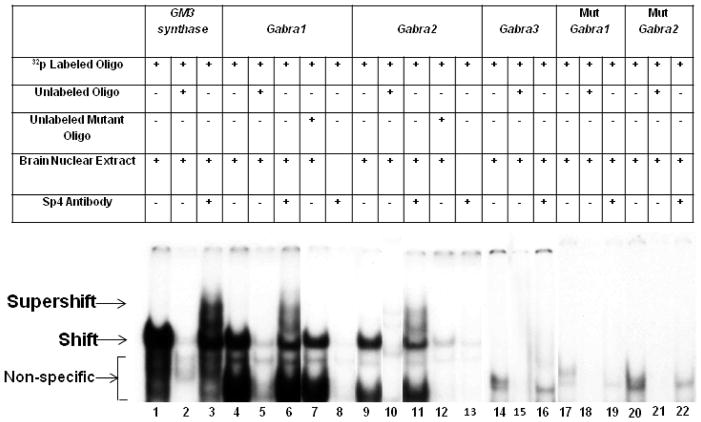
Fig. 1.
In vitro binding of Sp4 transcription factor to putative binding sites on GABAA receptor subunit promoters using EMSA and supershift assays. All lanes contain 32P-labeled oligonucleotides and are indicated by a “+” or a “−” sign depending on whether they also contain mouse brain nuclear extract, excess unlabeled oligos, unlabeled mutant oligos, or Sp4 antibody. Sp4 shift, supershift, and non-specific complexes are indicated by arrows. The positive control for Sp4 factor binding was GM3 Synthase. Specific Sp4 shift bands were revealed upon incubation with cortical nuclear extract (lane 1). Excess unlabeled competitor competed out the shift band (lane 2). The addition of Sp4 antibody yielded a strong supershift band (lane 3) corresponding to the presence of tandem Sp binding. Incubation of cortical nuclear extract with the Gabra1 and Gabra2 probes yielded specific Sp4 shift bands (lane 4 and 9, respectively) that were competed out with the addition of excess cold probes (lanes 5 and 10, respectively). The addition of Sp4 antibody yielded fainter but specific supershift bands (lanes 6 and 11, respectively). The addition of excess unlabeled mutant Gabra1 and Gabra2 probes did not compete out the shift reaction (lanes 7 and 12, respectively). Labeled Gabra1 and Gabra2 with Sp4 antibody without any nuclear extract was used as a positive control, to check for any antibody-oligo interaction, and they did not show any shift or supershift bands (lanes 8 and 13). Labeled Gabra3 probes did not yield specific Sp4 shift (lane 14) or supershift bands (lane 16), and cold competitor was also blank (lane 15). Labeled Gabra1 and Gabra2 probes with mutant Sp sites did not yield specific Sp4 shift or supershift bands (lanes 17–19 and 20–22, respectively).