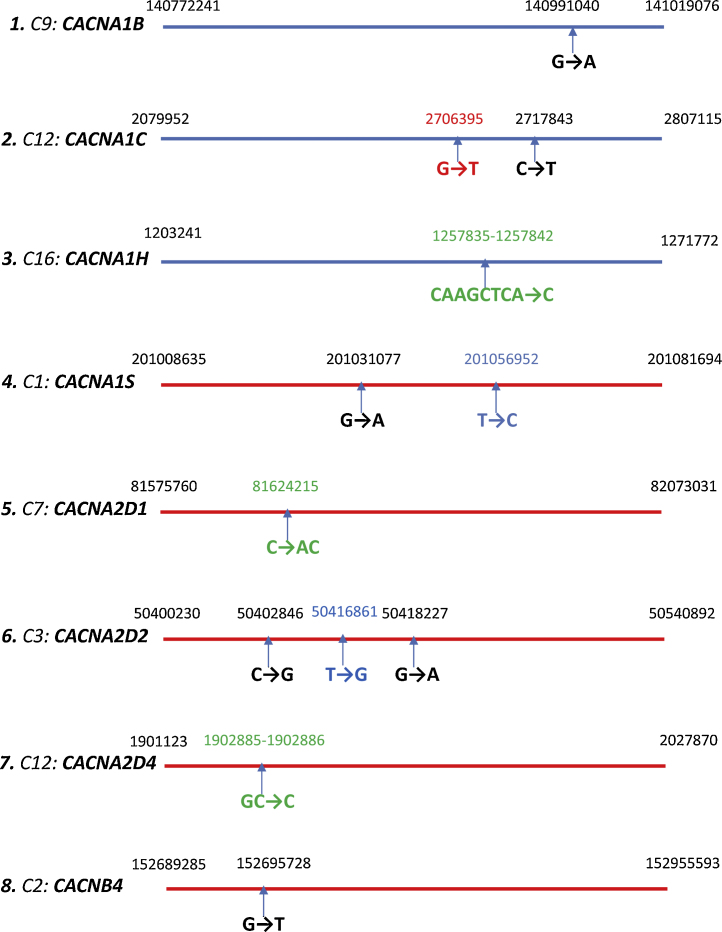
Mutations identified in exome sequencing. Analysis of mutations that are highly likely to be disruptive, identified in a whole exome sequencing of schizophrenia (
). The line in each case represents the appropriate full-length gene. The chromosome number is indicated next to the gene name. The beginning and end nucleotide position of each gene on the chromosome is shown (using GRCh 37). Nucleotide mutations are indicated with an arrow below the line and the chromosome nucleotide position is given above the line. The red lines correspond to proteins which are coded on the minus strand; however all of the nucleotides referred to are shown as coding strand for that particular gene. The code for the mutation sites is: red = splice acceptor mutation; blue = splice donor mutation; black = point mutation to stop codon; green = frameshift. Additional non-synonymous single nucleotide variants that change a single residue have not been included in the diagram but are listed in the Supplementary information.
-
1)
CACNA1B: G→A will convert a tryptophan residue into a stop codon, truncating the protein.
-
2)
CACNA1C: G→T is a splice acceptor mutation, whereas C→T will convert a glutamine residue into a stop codon, truncating the protein.
-
3)
CACNA1H: CAAGCTCA→C is a 7 nucleotide deletion, causing a frameshift leading to addition of 7 different amino acids followed by a stop codon—the original protein is 2347aa, this truncated protein is 1050aa. A deletion at position 1260920, TC→T, causes a frameshift leading to a truncation (15 missense aa before the stop codon-truncated protein is 1406aa, the WT protein is 2347aa).
-
4)
CACNA1S: G→A will convert a tryptophan residue into a stop codon, truncating the protein. T→C is a splice donor mutation.
-
5)
CACNA2D1: C→AC is an insertion, disrupting the amino acid code thereafter due to frameshift. It will cause truncation of the protein.
-
6)
CACNA2D2: C→G will convert a tyrosine residue into a stop codon, truncating the protein. T→G is a splice donor mutation. G→A will convert a tryptophan residue into a stop codon, truncating the protein.
-
7)
CACNA2D4: GC→C single nucleotide deletion causes a frameshift, changing the following 21 amino acids and adding an additional 28 amino acids before a stop codon occurs. The original protein is 1137aa, this predicted mutated protein is 1165aa.
-
8)