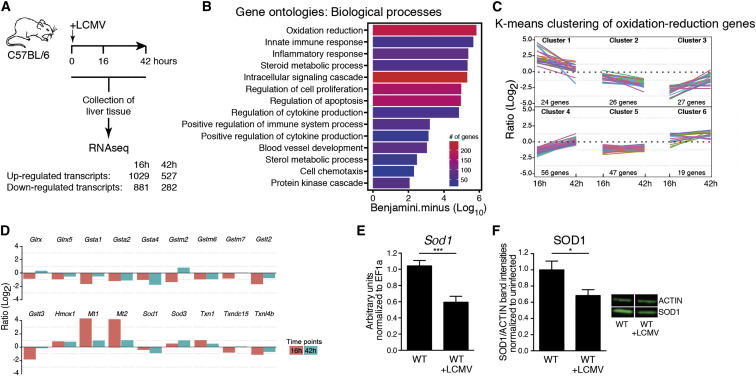
Figure 1.
Viral Infection Results in Transcriptional Regulation of Oxidation-Reduction Pathways in the Liver
(A–D) Wild-type (WT) mice were infected with LCMV. Profiling of liver tissue harvested at the indicated time points was performed by RNA-seq (n = 3 mice).
(A) Workflow and summary of up- and downregulated transcripts. See also Table S1.
(B) Gene ontology enrichment analysis of significantly regulated transcripts from RNaseq data.
(C) K-means clustering of gene regulatory profiles from the GO term oxidation reduction. Individual genes are represented by different colored lines. See also Table S2.
(D) Selection of differentially regulated genes involved in oxidation-reduction related processes (Experimental Procedures).
(E) mRNA expression of Sod1 was determined by real-time PCR from liver tissue of WT mice that were either left uninfected or infected with LCMV 44 hr previously (n = 7–12 mice per group pooled from three independent experiments).
(F) Western blot for SOD1 and actin were performed from liver lysates of WT mice that were either left uninfected or infected with LCMV 44 hr previously (representative results are shown). Relative protein ratios of SOD1 to actin were quantified by LI-COR (n = 11 mice per group pooled from three independent experiments). Statistical significance was calculated by unpaired t test (E and F). Symbols represent the mean ± SEM.