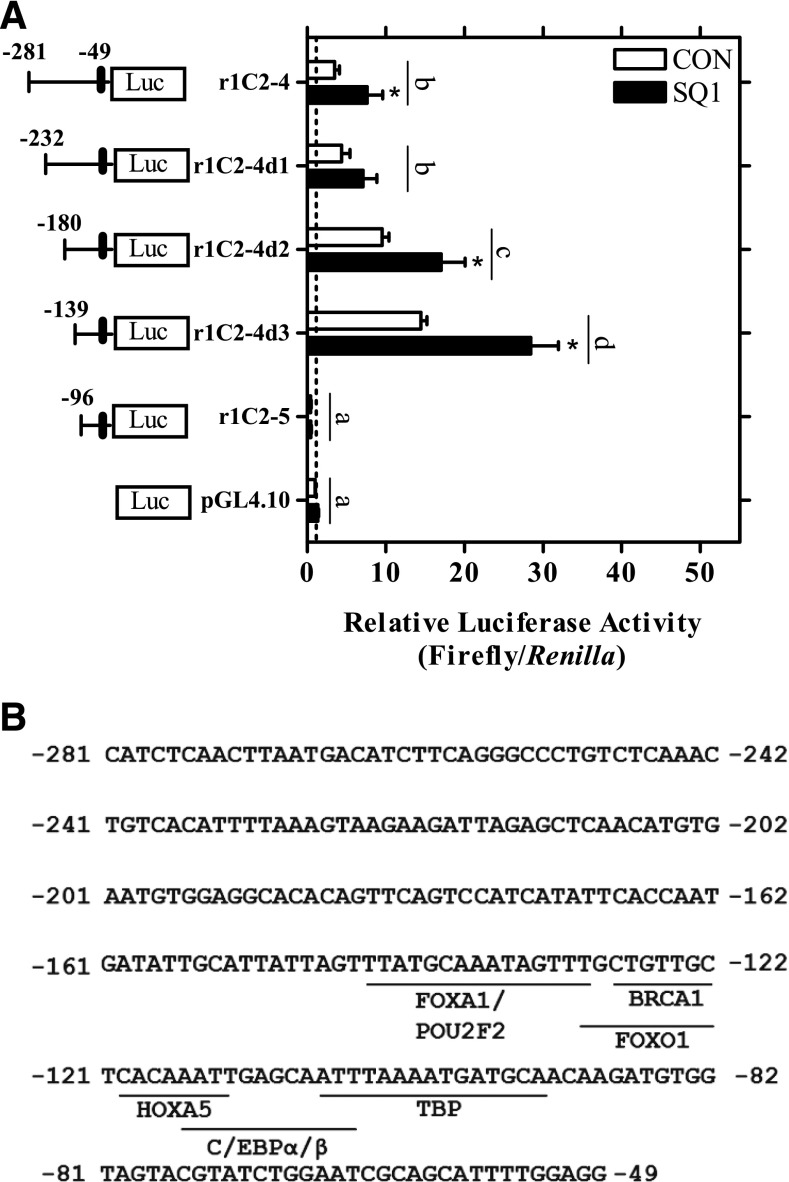
Fig. 7.
Evaluation of the SQ1-responsive region of the SULT1C2 promoter. (A) Three firefly luciferase deletion constructs based on the r1C2-4 sequence (-281:-49) were prepared. Twenty-four hours after plating, primary cultured rat hepatocytes were transfected with a SULT1C2 reporter plasmid (r1C2-4, deletion constructs r1C2-4d1-3, or r1C2-5) or with the empty vector as a control (pGL4.10). The next day, the hepatocytes were treated with Williams’ E medium alone (CON, control) or containing 0.1 µM SQ1. The medium containing the treatments was replaced once after 24 hours. Ninety-six hours after plating, cells were harvested for the measurement of luciferase activity. Bars represent mean ± S.E.M. of luciferase measurements (firefly/Renilla) normalized to untreated, empty vector controls. Results are combined from three independent experiments, with each independent experiment representing one rat hepatocyte preparation (n = 3 wells/treatment/experiment). *Statistically significant effect of SQ1 treatment within each reporter construct compared with the untreated, construct-matched control (CON) (P < 0.05). Different letters denote significant differences between the individual constructs on luciferase reporter activity (P < 0.05). (B) Transcription factor binding sites identified in the (-145:-49) promoter region predicted using the JASPAR database.