Abstract
Because lung cancer is the most common cause of cancer death among both men and women, focused efforts are necessary to identify and develop biomarkers that aid in the detection and treatment of this serious disease. Recent research has been aimed at understanding the roles of microRNAs (miRNAs) in tumorigenesis and their utility as cancer biomarkers. Here, miR-21 was investigated as a potential serum biomarker for non-small cell lung cancer (NSCLC). The relative expression level of miR-21 was detected by real-time PCR in the sera of 80 NSCLC patients; sera were also collected from 60 healthy people as a control. The most suitable cut-off value and the prognostic value of serum miR-21 levels were analyzed using a receiver-operating curve. The relative serum miR-21 level in NSCLC patients was significantly higher than that in healthy people (P<0.05). For relative miR-21 expression, the area under the ROC curve was 0.812 (95% CI: 0.736-0.888) with a sensitivity of 73.8% and a specificity of 71.7%, based on a cut-off value of 1.22. NSCLC patients were divided into two groups based on miR-21 expression; those with higher relative expression (≥1.22) had significantly lower survival time than those in the lower expression group (P<0.05). Further, serum miR-21 level and survival time were negatively correlated in NSCLC patients (P<0.05). Thus, miR-21 may be useful as a diagnostic and prognostic indicator of NSCLC.
Keywords: MiR-21, non-small cell lung cancer, serum, diagnosis, prognosis
Introduction
Lung cancer is the second-most common cancer and is the leading cause of cancer death worldwide [1]. Two pathological types of this disease exist: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC is more common, accounting for more than 80% of lung cancer patients [2]. Importantly, lung cancers often do not cause symptoms until the disease reaches an advanced stage [1]. This feature, along with high mortality, has prompted focused research efforts on identifying biomarkers to aid in the detection and treatment of lung cancer.
Recent efforts in cancer biology have sought to understand the roles of microRNAs (miRNAs) in tumorigenesis and treatment response. miRNAs are non-coding, short, single-stranded RNAs that regulate gene expression through the degradation, inhibition, or activation of target mRNA. Importantly, miRNAs have been associated with both the occurrence and development of various tumors [3-5]. miR-21 is considered to be a classic oncogene; indeed, its expression is positively correlated with cancer metastasis [6,7]. At the same time, miR-21 was also the first miRNA molecule detected in serum and, recently, its serum expression level has been linked with the prognosis of lung cancer patients [8,9]. This study sought to verify the value of detecting the serum expression level of miR-21 for the diagnosis and prognosis of NSCLC in Han Population in Henan Province, Central China.
Materials and methods
Participants
The study prospectively enrolled 80 NSCLC patients who were treated in our hospital from January 2012 to December 2013. The group comprised 49 males and 31 females, ages 44 to 71 (mean, 57.61±8.0) years. None of the patients had received chemotherapy or radiation therapy before enrollment. An additional 60 healthy individuals who underwent physical examination in our hospital during the same time period were included as the control group. This group comprised 31 males and 29 females, ages from 43 to 65 (mean, 55.4±6.6) years. There was no statistically significant difference between the patient group and control group in terms of gender or age (P<0.05). This study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, Henan Province, China, and all participants provided informed consent.
Research methods
Extraction of serum RNA
A total of 5 mL venous blood was collected from each fasting participant. Samples were centrifuged immediately after coagulation, and the supernatant was collected and stored at -80°C. Total RNA was extracted from serum using the Trizol method. RNA purity and concentration was determined with a UV spectrophotometer for absorbance (A) 260/280 value. Samples with a 1.8-2.0 value would be included in subsequent experiments.
Reverse transcription and quantitative real-time PCR
Extracted total RNA was reverse-transcribed according to the instructions of the reverse transcription kit (ABI, USA) to obtain cDNA. Primer Premier 5.0 software was used to design primers according to miR-21 and U6 gene sequences provided in GenBank. The primers, synthesized by ABI (USA), were as follows: miR-21 upstream, 5’-GTTAGCTTATCAGACTGA-3’; miR-21 downstream, 5’-GTGCAGGGTCCGAGGTAT-3’; U6 upstream, 5’-CTCGCTTCGGCAGCACA-3’; and U6 downstream, 5’-AACGCTTCACGAATTTGCGT-3’. The expected product size for miR-21 was 66 bp, for U6 was 94 bp. PCR was performed in a total volume of 20 μL comprising 10 μL TaqMan Gene Expression Master Mix (Life Technologies, USA); 0.5 μL of each primers (concentration: 10 μmol/L), 4 μL ddH2O. The reaction was performed in a thermal cycler as follows: pre-degeneration at 95°C for 10 minutes; and 40 cycles of 95°C for 12 s and 62°C for 50 s. Each sample was performed in triplicate. Fluorescence signal was measured; the relative expression of miR-21 was determined with 2-ΔΔCT value [10].
Follow-up
NSCLC patients were followed for 12 to 48 months through home visits, letters, and telephone once every 3 months. Information collected included whether the patients received reexaminations on schedule, as well as recurrence and metastasis time, and causes and times of death. All 80 patients were followed up completely.
Statistical methods
All test results were processed with SPSS17.0 (IBM) statistical package. An independent sample t-test was used to analyze the difference in serum miR-21 levels between the two groups. A receiver-operating characteristic (ROC) curve was established to evaluate the diagnostic value of serum miR-21 for NSCLC. A Kaplan-Meier survival analysis was performed, and Pearson correlation analysis was conducted between the serum miR-21 levels and survival time. For all tests, P<0.05 was considered as statistically significant.
Results
Serum detection of miR-21 expression
The average relative expression of miR-21 (Figure 1) in NSCLC patients (1.41±0.23) was significantly higher than that of control individuals (0.94±0.42) (t=8.376, P=0.001).
Figure 1.
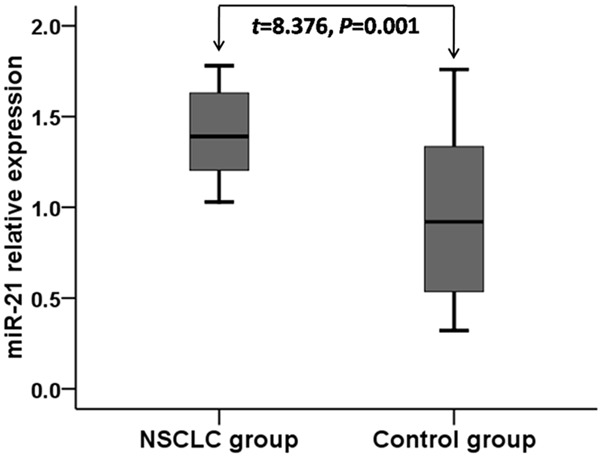
Relative expression of miR-21 in NSCLC patients and healthy controls.
Diagnostic value of miR-21 for NSCLC
An ROC curve was created to determine the diagnostic value of serum miR-21 expression levels. The area under the curve was 0.812 [95% confidence interval (CI): 0.736-0.888]. The sensitivity and specificity were 73.8% and 71.7%, respectively, when the cut-off value was 1.22, as shown in Figure 2.
Figure 2.
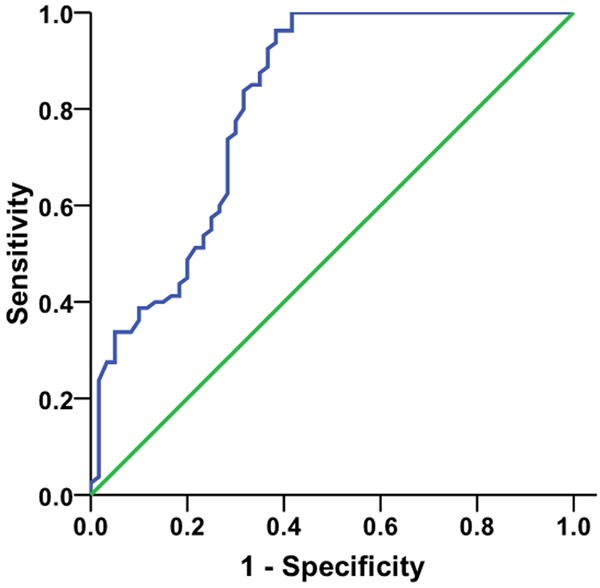
ROC curve for diagnostic value of serum miR-21 in NSCLC.
Relationship between serum miR-21 expression level and prognosis for NSCLC patients
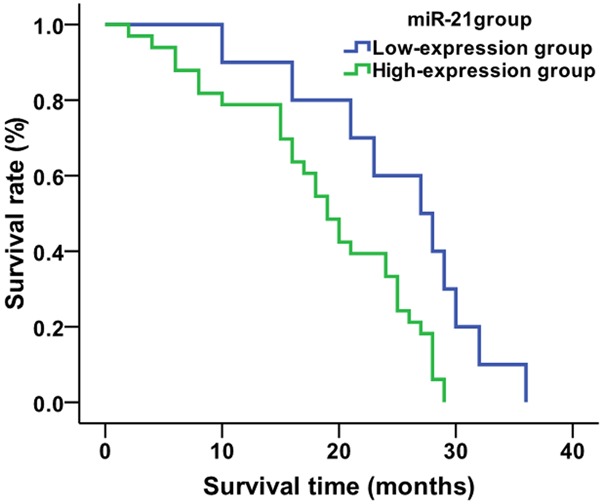
Using the average value (1.22) for relative expression of serum miR-21 in NSCLC patients as the cut-off value, the 80 NSCLC patients were divided into two groups: 21 patients in the miR-21 low-expression group (relative expression of miR-21<1.22, 11 patients accepting chemo/radio therapies and 10 not accepting chemo/radio therapies ) and 59 patients in the miR-21 high-expression group (relative expression ≥1.22, 26 treated patients and 33 untreated patients). Kaplan-Meier analysis showed that the average survival time of patients in the miR-21 low-expression group was 34.4 months, and that of miR-21 high-expression group was 23.1 months (Figure 3). The survival time of patients in the miR-21 low-expression group was significantly higher than that of the miR-21 high-expression group (χ2=16.409, P=0.001). When looking only at the treated patients in each expression group, the average survival times of patients who were treated in the miR-21 low-expression group and miR-21 high-expression group were 42.7 and 28.6 months, respectively (Figure 4). The survival time of treated patients in the miR-21 low-expression group was significantly higher compared to the miR-21 high-expression group (χ2=15.314, P=0.001). The average survival times of untreated patients in the miR-21 low-expression group and miR-21 high-expression group were 25.2 and 18.8 months, respectively (χ2=6.788, P=0.009) (Figure 5).
Figure 3.
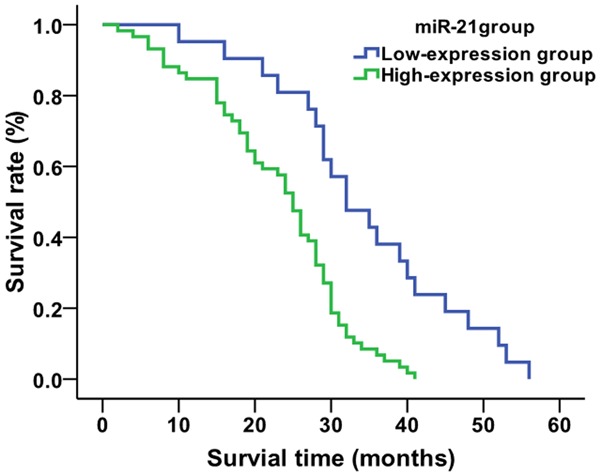
Comparison of average survival time of NSCLC patients based on miR-21 expression level.
Figure 4.
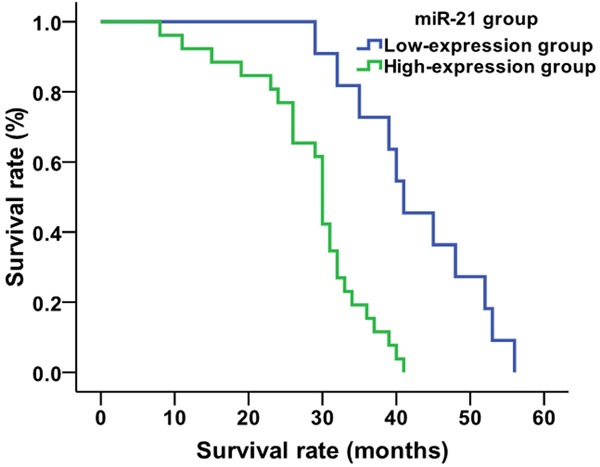
Comparison of average survival time of treated NSCLC patients based on miR-21 expression level.
Figure 5.
Comparison of average survival time of untreated NSCLC patients based on miR-21 expression level.
Relationship between serum miR-21 expression level and average survival times for NSCLC patients
A Pearson correlation analysis revealed a negative relationship between serum miR-21 level and survival time in NSCLC patients (r=-0.508, P=0.001, Figure 6). Similarly, when NSCLC patients were analyzed by treatment status, a negative relationship was detected between serum miR-21 level and average survival time of treated NSCLC patients (r=-0.701, P=0.001, Figure 7), as well as untreated NSCLC patients (r=-0.509, P=0.001, Figure 8).
Figure 6.
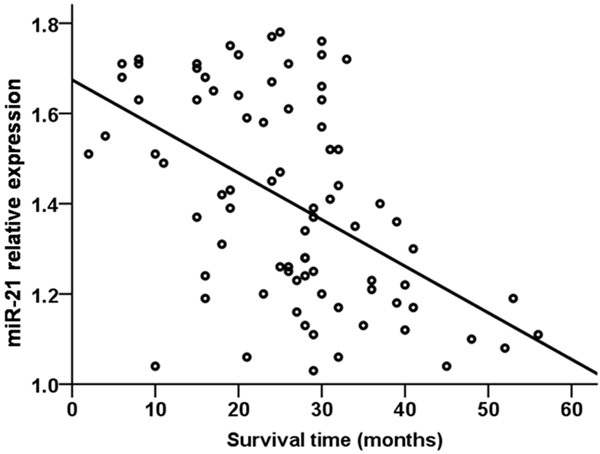
Relationship between serum miR-21 level and survival time in NSCLC patients.
Figure 7.
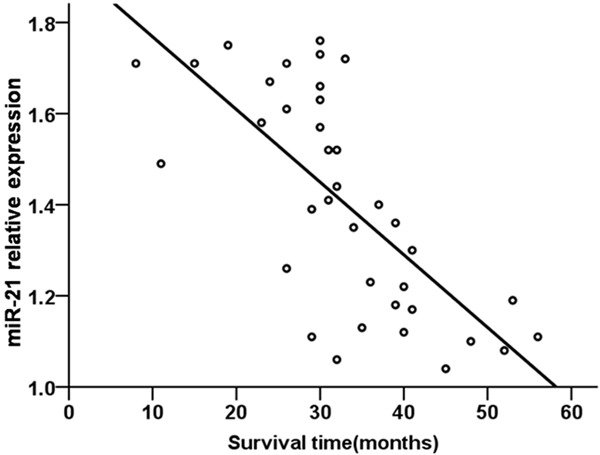
Relationship between serum miR-21 level and survival time in treated NSCLC patients.
Figure 8.
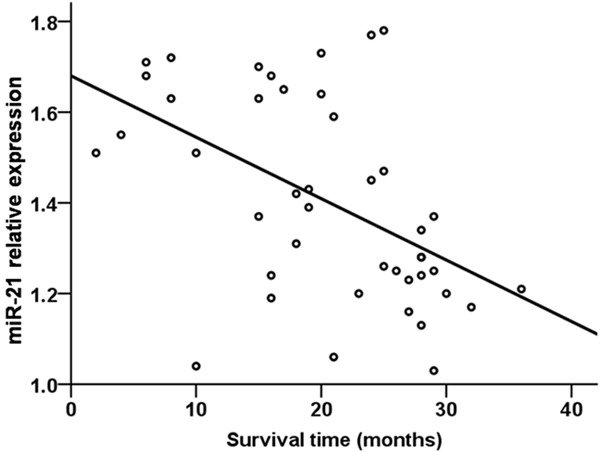
Relationship between serum miR-21 level and survival time in untreated NSCLC patients.
Discussion
Abnormal cell proliferation is an important feature of tumor biology, and miRNA plays an important regulatory role in cell proliferation, differentiation, and apoptosis [11]. Numerous studies have reported that miRNAs act as oncogenes or cancer suppressor genes; some miRNAs with oncogenic effects can promote tumorigenesis by inhibiting expression of tumor suppressor genes [12]. The expression level of miRNAs in tumor tissues differs significantly from that of normal tissues [13,14], thus these abnormally expressed miRNA are expected to be important markers for the diagnosis, prognosis, and treatment of tumors.
miR-21 expression is significantly associated with tumorigenesis, displaying increased expression in breast, liver, lung, and gastric cancers [15,16]. Further, the inhibition of miR-21 expression can promote tumor apoptosis [17]. In this study, quantitative PCR was used to detect serum miR-21 expression in NSCLC patients and healthy individuals to determine its potential utility as a biomarker of NSCLC. Indeed, miR-21 expression was significantly higher in the serum of NSCLC patients than healthy indiviuals, indicating that serum miR-21 expression may reflect the occurrence and development of NSCLC. The sensitivity (73.8%) and specificity (71.7%) of detecting miR-21 expression in the serum further confirmed that serum miR-21 level can be used as a potential tumor marker for diagnosis of NSCLC. In addition, a survival analysis correlated the expression of serum miR-21 with survival of patients with NSCLC; lower serum miR-21 expression was associated with longer survival time compared to patients with higher serum miR-21 expression. Pearson correlation analysis showed there were negative relationships between serum miR-21 level and survival time in NSCLC patients; specifically, the survival time decreased with increasing serum miR-21 level. Thus, serum miR-21 expression levels may be useful as a diagnostic and prognostic indicator for NSCLC.
In summary, serum miR-21 expression is higher in NSCLC patients and higher miR-21 expression in the serum is associated with shorter survival. Thus, miR-21 may be useful as a primary molecular marker for the diagnosis and prognosis of NSCLC. However, its clinical application value still needs to be confirmed in further research.
Disclosure of conflict of interest
None.
References
- 1.Cersosimo RJ. Lung cancer: a review. Am J Health Syst Pharm. 2002;59:611–642. doi: 10.1093/ajhp/59.7.611. [DOI] [PubMed] [Google Scholar]
- 2.Rezaei MK, Nolan NJ, Schwartz AM. Surgical pathology of lung cancer. Semin Respir Crit Care Med. 2013;34:770–786. doi: 10.1055/s-0033-1358558. [DOI] [PubMed] [Google Scholar]
- 3.Du L, Pertsemlidis A. Cancer and neurodegenerative disorders: pathogenic convergence through microRNA regulation. J Mol Cell Biol. 2011;3:176–180. doi: 10.1093/jmcb/mjq058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan B, Manley J, Lee J, Singh SR. The emerging roles of microRNAs in cancer metabolism. Cancer Lett. 2015;356:301–308. doi: 10.1016/j.canlet.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Ibrahim SA, Hassan H, Götte M. MicroRNA regulation of proteoglycan function in cancer. FEBS J. 2014;281:5009–5022. doi: 10.1111/febs.13026. [DOI] [PubMed] [Google Scholar]
- 6.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Yang YB, Zhang XH, Yu XL, Wang ZB, Cheng XC. MicroRNA-21 gene and cancer. Med Oncol. 2013;30:376. doi: 10.1007/s12032-012-0376-8. [DOI] [PubMed] [Google Scholar]
- 8.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 9.Heegaard NH, Schetter AJ, Welsh JA, Yoneda M, Bowman ED, Harris CC. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int J Cancer. 2012;130:1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liva KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Bishop JA, Benjamin H, Cholakh H, Chajut A, Clark DP, Westra WH. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin Cancer Res. 2010;16:610–619. doi: 10.1158/1078-0432.CCR-09-2638. [DOI] [PubMed] [Google Scholar]
- 14.Corsini LR, Bronte G, Terrasi M, Amodeo V, Fanale D, Fiorentino E, Cicero G, Bazan V, Russo A. The role of microRNAs in cancer: diagnostic and prognostic biomarkers and targets of therapies. Expert Opin Ther Targets. 2012;(Suppl 2):S103–109. doi: 10.1517/14728222.2011.650632. [DOI] [PubMed] [Google Scholar]
- 15.Huang GL, Zhang XH, Guo GL, Huang KT, Yang KY, Shen X, You J, Hu XQ. Clinical significance of miR-21 expression in breast cancer: SYBRGreen I-based real-time RT-PCR study of invasive ductal carcinoma. Oncol Rep. 2009;21:673–9. [PubMed] [Google Scholar]
- 16.Huang Y, Yang YB, Zhang XH, Yu XL, Wang ZB, Cheng XC. MicroRNA-21 gene and cancer. Med Oncol. 2013;30:376. doi: 10.1007/s12032-012-0376-8. [DOI] [PubMed] [Google Scholar]
- 17.Cui L, Zhang X, Ye G, Zheng T, Song H, Deng H, Xiao B, Xia T, Yu X, Le Y, Guo J. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer. 2013;119:1618–1626. doi: 10.1002/cncr.27903. [DOI] [PubMed] [Google Scholar]


