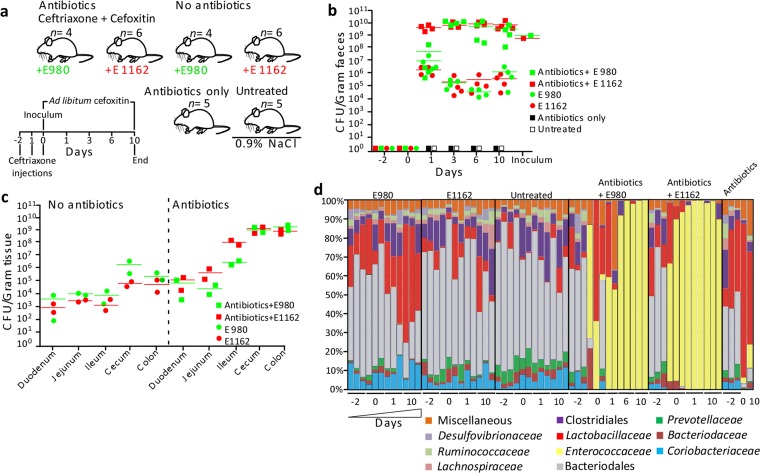
FIG 1 .
E. faecium colonization levels and 16S rRNA gene-based microbiota profiling. (a) Schematic overview of the animal model used in this study. Mice were treated with the antibiotics ceftriaxone and cefoxitin and inoculated with E. faecium E980 (n = 4) and E1162 (n = 6) on day 0 or treated with 0.9% NaCl and inoculated with E980 (n = 4) and E1162 (n = 6). Control mice were treated with the antibiotics only (n = 5) or left untreated (n = 5). (b, c) Quantification of E. faecium CFU in mouse feces (n = 22) (b) and in homogenized small and large intestinal parts (c) on Slanetz-Bartley agar (two mice per group). (d) Percentages of 16S rRNA gene sequences assigned to taxonomic bacterial genera present in feces during the intestinal colonization experiment. Each bar represents one mouse on 1 sampling day, and different taxa are indicated by different colors. Fecal pellets of three mice per time point were used for 16S rRNA gene sequencing. However, in some instances, not enough DNA could be extracted or the sequencing run failed. In those cases, data based on fecal pellets of only two mice per time point are shown. This was the case for antibiotic-treated mice inoculated with E980 on days 1 and 10 and mice treated with the antibiotics only on days 0, 1, and 10. For mice treated with the antibiotics and inoculated with E. faecium E980, an additional time point, day 6, was analyzed to compensate for the missing day 1 and 10 data.