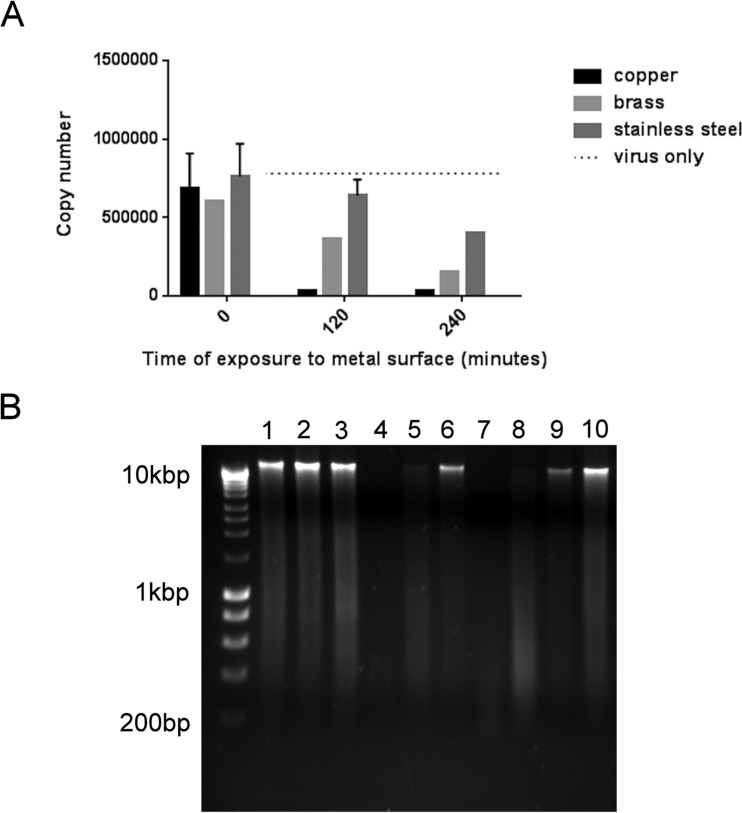
FIG 5 .
Destruction of human coronavirus viral genome on copper and copper alloy surfaces. (A) Analysis of a small fragment (136-bp region of the nsp4 gene) of the coronavirus genome revealed a reduction in copy number from virus exposed to copper and cartridge brass surfaces in reverse transcriptase real-time PCR. There was some reduction on stainless steel but none in viral suspension (lightest gray bars), suggesting that this was due to sample drying. (B) Analysis of the entire viral genome is represented in electrophoretic separation of viral RNA extracted from virus exposed to copper (lanes 1, 4, and 7), cartridge brass (lanes 2, 5, and 8), and stainless steel (lanes 3, 6, and 9) for 0 min (lanes 1 to 3), 120 min (lanes 4 to 6), and 240 min (lanes 7 to 9). The genomic RNA from virus exposed to copper and brass degraded with increased contact time. This did not occur on stainless steel; the genomic RNA remained as fragments too large to pass through the gel. However, the total amount of intact RNA was reduced at 4 h, possibly due to drying damage as seen in panel A. Lane 10 represents untreated virus, and the unmarked lane is a Bioline marker (Hyperladder I). The same procedure was used with mock-infected cells, revealing the same pattern of RNA breakdown following application to copper surfaces (not shown).