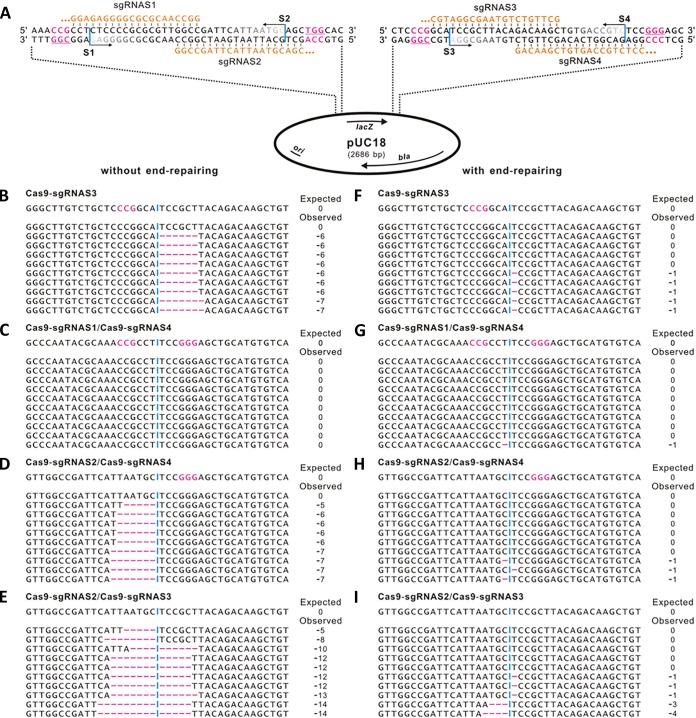
FIG 1 .
Recombination of a Cas9-created DNA fragment with and without end repair. (A) Schematic representation of four selected protospacers in the pUC18 plasmid. The transcribed sgRNAs matched the specific protospacers are shown in orange. PAM sequences are highlighted in pink, and “NGG” is underlined. The vertical blue lines between nucleotides indicate the Cas9-mediated DSB sites matched with specific protospacers. The 3′→5′ exonuclease trimming is shown with horizontal arrows, and the sequential trimming occurred in the noncomplementary strand is shown in gradient gray. ori, origin of replication of plasmid pUC18; bla, ampicillin resistance gene; lacZ, β-galactosidase gene. (B to E) End-joint sequencing results from cleavage of Cas9-sgRNAS3 (B), Cas9-sgRNAS1/Cas9-sgRNAS4 (C), Cas9-sgRNAS2/Cas9-sgRNAS4 (D), and Cas9-sgRNAS2/Cas9-sgRNAS3 (E) without end repair. For each combination of Cas9-sgRNA complex, the desired sequence is shown at the top, with the PAM sequence highlighted in pink and the joint interface indicated with a vertical blue line. Deletions are shown as a pink dashed line. The net change in length caused by exonuclease trimming is shown to the right of each sequence (−, deletion). (F to I) End-joint sequencing results from cleavage of Cas9-sgRNAS3 (F), Cas9-sgRNAS1/Cas9-sgRNAS4 (G), Cas9-sgRNAS2/Cas9-sgRNAS4 (H), and Cas9-sgRNAS2/Cas9-sgRNAS3 (I) with an additional end-repairing procedure that is parallel to the left column, respectively.