Fig. 7.
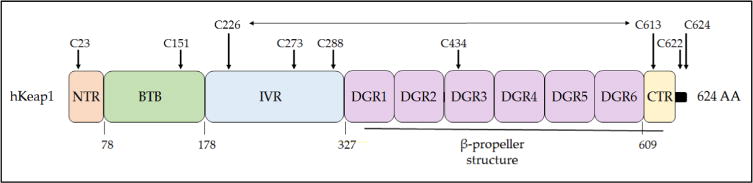
Domain structure of human Keap1. The Keap1 protein can be divided into five domains, the N- and C-terminal NTR and CTR sequences and the BTB, IVR, and DGR sequences. The BTB domain is responsible for dimerization and also the recruitment of cullin-3 to the CRLKeap1 complex. The DGR domain along with the CTR form the β-propeller structure to which Nrf2 binds, with the DLG motif in the Neh2 domain of Nrf2 docking onto one Keap1 subunit and the ETGE motif docking onto the other Keap1 subunit. Cysteine residues that are functionally important or unusual are highlighted: Cys-151 is crucial for the ability of tBHQ and SFN to inhibit the substrate adaptor activity of Keap1 [250], and it forms a transient disulfide bridge with Cys-151 in the other Keap1 subunit [251]; Cys-273 and Cys-288 recognize alkenals and cyclopentanone prostaglandins [252, 253]; Cys-434 recognizes 8-nitro-cGMP [254]; Cys-226 and Cys-613 recognize metals such as Zn2+, Cd2+, As3+, and Se4+[253], and they form a transient disulfide bridge upon exposure to H2O2 [113, 251], which is represented by a horizontal two-headed arrow. Mutation of Cys-23 to tyrosine has been shown to impair the ability of Keap1 to ubiquitylate Nrf2 in breast cancer cells [255, 256]. The three amino acids at the C-terminus of human, mouse, and rat Keap1 comprises a distinctive CTC motif but its functional significance is obscure.
