Abstract
CD4+ T cells are critical in the fight against parasitic, bacterial, and viral infections, but are also involved in many autoimmune and pathological disorders. Studies of protein function in human T cells are confined to techniques such as RNAi due to ethical reasons and relative simplicity of these methods. However, introduction of RNAi or genes into primary human T cells is often hampered by toxic effects from transfection or transduction methods that yield cell numbers inadequate for downstream assays. Additionally, the efficiency of recombinant DNA expression is frequently low due to multiple factors including efficacy of the method and strength of the targeting RNAs. Here, we describe detailed protocols that will aid in the study of primary human CD4+ T cells. First, we describe a method for development of effective microRNA/shRNAs using available online algorithms. Second, we illustrate an optimized protocol for high efficacy retroviral or lentiviral transduction of human T cell lines. Importantly, we demonstrate that activated primary human CD4+ T cells can be transduced efficiently with lentiviruses, with a highly activated population of T cells receiving the largest number of copies of integrated DNA. We also illustrate a method for efficient lentiviral transduction of hard-to-transduce un-activated primary human CD4+ T cells. These protocols will significantly assist in understanding the activation and function of human T cells and will ultimately aid in the development or improvement of current drugs that target human CD4+ T cells.
Keywords: Human CD4+ T cells, Lentiviruses, ShRNA development, MicroRNA, Transductions
INTRODUCTION
Alteration of the overall transcriptome and proteome of T cells through multiple methods is an important approach in elucidating the mechanisms of T cell activation and their role in human disease. A considerable amount of work has utilized gene knockout (KO) technology in understanding T cell biology and development1, 2, 3. Due to technical and ethical reasons, these studies are limited to mice and other research animals. However, due to complexity of human disease, it is becoming increasingly important to translate research derived from murine studies to more relevant human models. This is especially critical since the biology of humans and mice is subtly different both in anatomy and cellular biology4, 5. Therefore, methods must be developed to manipulate the genome of human immune cells.
Various techniques have been developed with the goal of inserting foreign nucleotides into human T cells. Several of these methods produce transient transfection of DNA including lipofection, polyethylene glycol, calcium phosphate, and electroporation. These methods vary in transfection efficiency based on a number of factors6, 7. In addition, some of these techniques, although efficient, yield significant toxicity and cell death depending on duration of transfection and the type of cells8. Production of stable lines expressing modified genes can be produced via transfections, but more rapidly and efficiently through retroviral-mediated stable integration7. Thus far, electroporation and retroviral transduction produce the highest efficiency of DNA delivery into human T cells6, 9, 10.
Retroviruses are RNA-based viruses that possess the ability to reverse transcribe their sequences into DNA, which is ultimately integrated into the host genome11. Due to their ability to efficiently infect a wide range of mammalian cells with relatively low toxicity, retroviruses have been used for gene therapy clinical trials for the treatment of human disease including inherited diseases and cancer11, 12, 13. Some of the most extensively utilized retroviruses are the murine-based gamma retroviruses Molony leukemia virus (MMLV) and stem cell virus (MSCV)14. HIV1-based lentiviruses, a subtype of retroviruses, are also widely utilized and are succeeding gamma retroviruses due to their ability to infect non-dividing and hard to infect cells11. To produce infectious viral particles, DNA of interest is cloned into a plasmid containing the viral genome and promoters recognized in mammalian cells such as the LTR, hU6, CMV and other promoters. The viral plasmid is then transiently co-transfected with vectors containing a viral envelope gene (i.e. VSV-G) and packaging sequences into packaging cells that produce pseudo-typed infectious retroviruses within hours. Due to separation of vectors containing viral genome, envelope, and packaging sequences, the viral particles are rendered replication incompetent once genomic integration is established11.
Due to their efficiency, low toxicity, simplicity of production, and relative safety, we have utilized retroviral transduction for the genomic modification of human CD4+ T cells. These methods have been successfully applied in recent studies utilizing human T cell lines as well as primary human CD4+ T cells15, 16. In this study, we describe in detail the optimized methods for the transduction of human primary CD4+ T cells and human T cell lines. Specifically, this manuscript includes specific details for producing retroviral particles carrying interference RNAs and reconstitution of mutated proteins. Importantly, we describe previously unreported specific conditions for high efficiency lentiviral transduction of human T cell lines, pre-activated primary human CD4+ T cells, and hard-to-transduce antigen-inexperienced primary human CD4+ T cells. We also present novel findings demonstrating that highly activated populations of T cells receive the largest number of copies of integrated DNA. Finally, we compare and contrast methods presented in this paper to current techniques utilized in modification of human primary T cells
RESULTS
Development of microRNAs and shRNAs
MicroRNAs are short non-coding RNA interference molecules (RNAi) produced in cells for the regulation and suppression of protein coding mRNAs. Due to their ability to suppress protein expression, RNAi have been implemented extensively in the analysis of gene function17, 18, 19. Upon transcription of RNAi, a hair-pin loop is formed that is processed and exported to the cytoplasm. The hair-pin loop is processed further into siRNA by Dicer and subsequently loaded into the RNA-induced silencing complex (RISC) complex targeting mRNAs specific to the loaded siRNA. The latter event leads to mRNA degradation or translation inhibition that results in protein suppression18, 20. Traditionally, shRNAs were utilized as an artificial suppression system due to the powerful protein expression inhibition effects. However, microRNA based mir-flanked shRNAs that mimic intrinsic RNAi-mediated suppression are predicated to have less toxicity or side effects and are shown to frequently produce increased inhibition levels17, 21, 22. Accordingly, we utilized mir30-based microRNAs as a primary suppression method and shRNAs in cases where the use of microRNAs is not suitable.
Development of suppressive microRNA sequences requires application of a few important guidelines. First, the majority of the microRNA core sequence must match the target mRNA. Second, the sequence must not have the potential to bind other target mRNAs; this is especially important for homologous proteins. Third, efficient suppression requires implementation of several nucleotide-based rules which may be applied alone or in combination for optimal inhibition18, 19. Because manual design of microRNAs is time consuming, difficult and inefficient, several online-based algorithms have been developed to mitigate this process by incorporating these rules into interactive software. In particular, we have found that the siRNA/shRNA microRNA-generating algorithm formulated by Dr. Sachidanandam and coworkers was highly successful in the suppression of several proteins (http://katahdin.cshl.edu). These sequences, although microRNA-based, may be used as shRNAs by removing the flanking mir30 sequences. In general, 10-40% of the generated sequences will produce 60-95% suppression efficiency. Occasionally, up to 15 sequences may need to be screened to find a sequence that achieves >90% protein inhibition.
Electroporation-mediated plasmid DNA delivery
Electroporation is one of the most effective methods for the introduction of DNA into human T cells. The main drawback of this method is the reduced cell viability and phenotypic changes6, 8, 23. Additionally, electroporated cells, particularly primary T cells, only transiently express delivered sequences; therefore this method is not as convenient compared to the development of stable cell lines via viral transductions. Nonetheless, in some cases gene delivery could be as efficient as retroviral-mediated transductions. Several instruments are available for the electroporation of human T cells. In particular, the square-wave pulse-based methods utilized by the Lonza Nucleofactor Amaxa electroporation system demonstrated high efficiency in gene delivery to T cells24. The high-cost of Lonza kits prompted some researchers to develop in-house electroporation buffers that are comparable to Lonza-based reagents6.
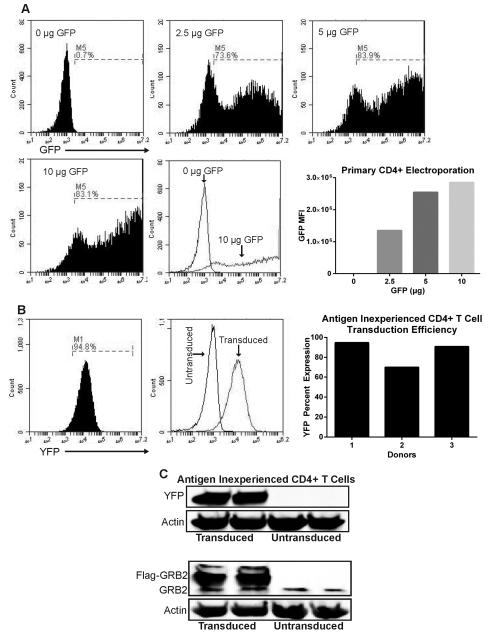
To test the efficiency of gene delivery via electroporation, primary CD4+ T cells were activated with magnetic CD3/CD28 beads and IL-2 for 3 days and then magnetic beads/IL-2 were removed from the culture. 5 × 106 activated primary CD4+ T cells were electroporated using reagents reported by Chicaybam et al (Reagent 1M)6. We found that this method could produce >80% transfection efficiency as measured by GFP expression greater than the non-transfected control (Figure 1a). Interestingly, there appeared to be three populations of cells upon transfection, an untransfected GFP- population that had overlapping GFP fluorescence with the non-transfected control, a GFP dim population with slightly increased fluorescence over the untransfected controls, and a GFP bright population with high expression of GFP. Moreover, there was a concentration dependent increase of mean GFP fluorescence and number of cells in the GFP bright population that peaks at 10 µg of plasmid DNA per 5 × 106 CD4+ T cells (Figure 1a). For obtaining cells with high-copy gene number, transfected CD4+ T cells can be sorted to enrich for CD4+ T cells expressing high copies of protein cDNA or shRNA.
Figure 1. Electroporation of activated T cells and transduction of antigen inexperienced primary human CD4+ T cells.
(A) Activated primary human CD4+ T cells were electroporated with various concentrations of pmaxGFP plasmid using Lonza Amaxa electroporator, and cells expressing GFP were detected after 24 hrs. Bottom right: Quantification of GFP MFI on electroporated cells. Data represents 3 independent experiments with similar results (B) Left: antigen inexperienced primary CD4+ were transduced with lentiviruses expressing LUC shRNA for 4 days and then YFP expression was measured. Right: Percent YFP expression of transduced antigen inexperienced CD4+ T cells compiled from three donors. (C) Antigen inexperienced primary CD4+ T cells were transduced for 4 days with lentiviruses carrying YFP or Flag-GRB2 constructs. Cells were lysed and protein expression of YFP, GRB2, and actin were detected via immunoblotting. Data represents 3 independent replicates with similar results.
Although this electroporation protocol generates high efficacy of gene input, the viability of electroporated T cells after 24 hrs was only in the range of 15-40% (data not shown). Our cell death observations are very similar to other T cell electroporation studies6, 23. Additionally, cell death continued to progress 2-3 days post electroporation and correlated with increased amount of plasmid DNA used. Therefore, we found that this method is not optimal for assays requiring high cell numbers (i.e. immunoprecipitations, signaling assays). Interestingly, in addition to electroporation-induced cell death, a recent study by Zhang et al. demonstrated multiple effects of electroporation on primary human CD4+ T cells23. Electroporated cells experience marked morphological changes before undergoing recovery after 1 hr. These cells had increased intracellular calcium levels for 8hrs and enhanced T cell activation markers (CD69, CD154) for more than 24 hrs23. Therefore caution must be taken if calcium assays and/or activation markers are to be analyzed shortly after CD4+ T cell electroporation. Due to the above reasons, we have focused our studies upon retro/lentiviral mediated DNA delivery15, 16.
Transduction of antigen inexperienced primary human CD4+ T cells
Lentiviral gene delivery to primary T cells in the absence of TCR activation is more challenging compared to primary T cells activated through TCR and CD28 pathways. The main reason for this appears to be the reduced proliferative and metabolic status in cells that were not activated in vitro. This results in a decrease in transduction efficiency and the number of viral copies present in each cell. However, previous studies reported the possibility of primary T cell transduction in the presence of cytokine stimulation9, 10. To this end, we tested whether our transduction protocols along with IL-2 treatment enhanced transduction efficiency of antigen inexperienced primary human CD4+ T cells.
We recommend centrifuge-mediated concentration of viral particles for all transductions. Transduction of primary CD4+ T cells with concentrated viral particles can have a significant effect on the efficiency of gene delivery when compared to dilute viral preparations. Nonetheless, this does not apply for cell lines as they are relatively easier to transduce with unconcentrated 293T viral supernatant. Next, there are reports of reagents which could significantly increase retroviral titer such as caffeine and sodium butyrate25, 26. However, one must be cautious of stimulatory or toxic side effects of these reagents which may carryover from viral preparation. Alternative methods of viral concentration are available (Polyethelene glycol, and commercial viral concentration solutions). However, these methods are not as cost-effective and also precipitate components of the media when compared to high speed ultra-centrifugation.
To this end, primary human CD4+ T cells were purified and the cells were treated with IL-2 and then transduced with lentiviruses expressing YFP. When assessed via flow cytometry, YFP was expressed on the majority of CD4+ T cells (Figure 1b). The YFP fluorescence corresponded to the GFP dim population observed in Figure 1a. This suggests that IL-2 stimulated T cells have a much lower capacity for high expression levels of transduced proteins. We found that YFP expression in different transductions was between 70-94% (Figure 1b). Importantly, these cells displayed significant expression of YFP protein via immunoblotting (Figure 1c). Moreover, antigen inexperienced CD4+ T cells could successfully be transduced with other lentiviral constructs. Specifically, antigen inexperienced cells were transduced with lentiviruses carrying Flag-GRB2 as illustrated via immunoblotting (Figure 1c). We have probed YFP expression for up to 6 days post initial transduction and observed minimal reduction of YFP expression (94% to 91% YFP expressing cells – data not shown) via flow cytometry. These data demonstrate that antigen inexperienced primary CD4+ T cells can be transduced efficiently with lentiviral constructs carrying the gene/protein of interest. However, due to low expression of viral products this method may not be effective for studies examining protein suppression effects via RNAi, but it appears suitable for protein expression studies.
Transductions, inhibition and reconstitution of shRNA-resistant proteins in human T cells
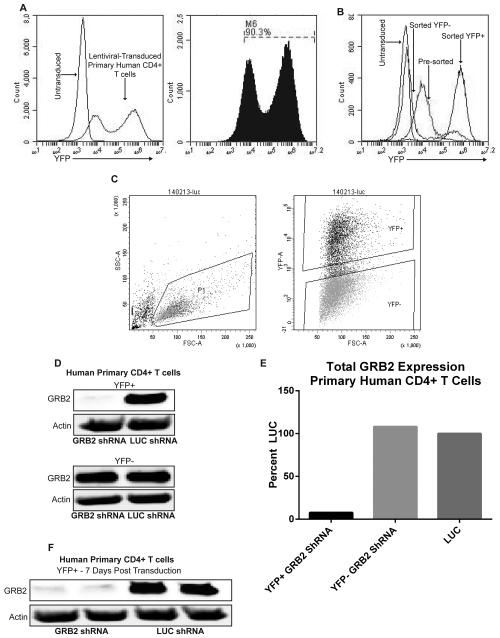
We next examined the viral transduction of antigen experienced human CD4+ T cells. Transduction efficiency of primary human CD4+ T cells ranges between 30-97% depending on the donor and viral preparation (Figure 2a). As seen previously in Figure 1a, we observed that there were three populations of YFP expression, YFP−, YFP dim and YFP bright. Puromycin may be used at this point to enrich cells containing viral constructs. These cells may also be used for protein studies. However this may not be optimal for shRNA-based studies due to the short lifetime of primary CD4+ T cells. To overcome this caveat, transduced primary CD4+ YFP bright T cells were sorted using a Becton Dickinson Aria II flow cytometer to obtain a population (>98%) of cells expressing specific shRNAs (Figure 2b and c). As illustrated, GRB2 expression in primary human CD4+ T cells was effectively suppressed by more than 90% after sorting of lentiviral-transduced YFP bright cells 3 days post transduction (Figure 2d and e). Importantly, GRB2 expression was still suppressed in primary human CD4+ YFP bright T cells 7 days post-transduction without the use of puromycin (Figure 2f).
Figure 2. Transduction of activated primary human CD4+ T cells using lentiviruses.
(A) Flow cytometry analysis of activated primary human CD4+ T cells that were transduced for 3 days with pLK4A lentiviral vectors carrying Luciferase (LUC) shRNA and YFP. Data represents 4 independent experiments with similar results. (B) Primary human CD4+ T cells were transduced with pLK4A lentiviral vectors expressing YFP along with LUC shRNA; illustrated groups are: untransduced, 3 day transduction pre-sort, Post-Sort YFP+, Post-Sort YFP−/low. (C) Left: Gate on live primary human CD4+ T cells transduced with pLK4A expressing YFP and LUC shRNA. Right: Gates for sorting YFP+ and YFP− populations. Data represents 3 independent replicates with similar results. (D) Primary human CD4+ T cells transduced with shRNAs against GRB2 or LUC were sorted into YFP+ and YFP− fractions after 3 days of transduction, lysed and probed for GRB2 expression. (E) Quantification of immunoblots from “D”. (F) Immunoblot of GRB2 expression from “D” 7 days post transduction.
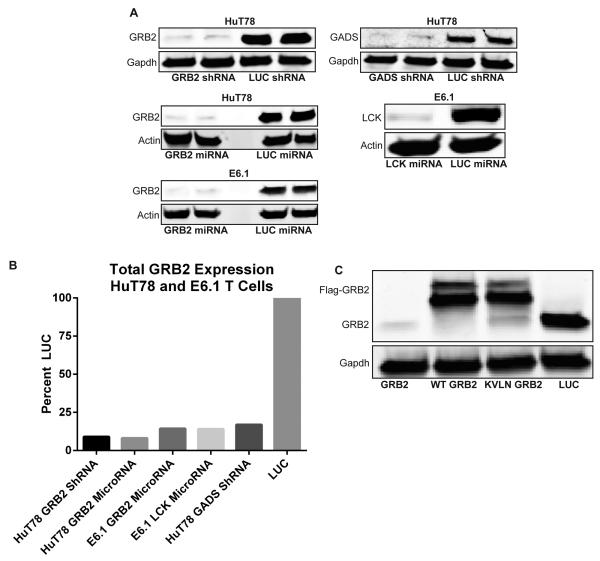
Next, stable T cell lines were produced via puromycin selection as described in materials and methods. GRB2 and GADS expression was effectively suppressed in lentiviral/retroviral transduced HuT78 and Jurkat T cells carrying microRNAs or shRNAs, respectively (Figure 3a and b). The most efficacious sequences produced more than 90% suppression of GRB2 and GADS in both HuT78 and E6.1 T cells. We were also able to suppress other proteins, including LCK, using the same microRNA/shRNA generating algorithm and transduction methods (Figure 3a and b). In contrast to electroporation-mediated protein suppression, protein suppression effects in our T cell lines are stable, as we have tested RNAi-mediated suppression after several months under moderate antibiotic selection and found no changes in protein suppression efficiency (data not shown). Finally, we utilized pLK4A lentiviruses to reconstitute suppressed protein expression using a Flag tagged, shRNA-resistant form of the protein (Figure 3c). The expression of reconstituted proteins is also constant in long term passages of the T cell lines. Overall, we demonstrate that retroviral mediated delivery of RNAi and simultaneous add-back of mutated proteins can be effectively used to study signal transduction in human T cells.
Figure 3. Lenti/retroviral-mediated inhibition of protein expression in human T cell lines.
(A) HuT78 or E6.1 “Jurkat” T cells were transduced with lentiviruses “hU6-shRNA” or retroviruses “LTR-microRNA” carrying hairpins against various proteins. Stable cell lines were lysed and probed for GRB2, GADS, and LCK via Immunoblotting. (B) Quantification of immunoblots from “A”. (C) HuT78 T cells were transduced with lentiviruses carrying LUC shRNA, and GRB2 shRNA with or without add-back of Flag-tagged shRNA-resistant wild-type or mutant GRB2 . Cells were lysed and proteins were probed with GRB2 and GAPDH antibodies. Data represents 3 independent replicates with similar results.
Differential efficiency of LTR and hU6 in RNAi-mediated protein suppression
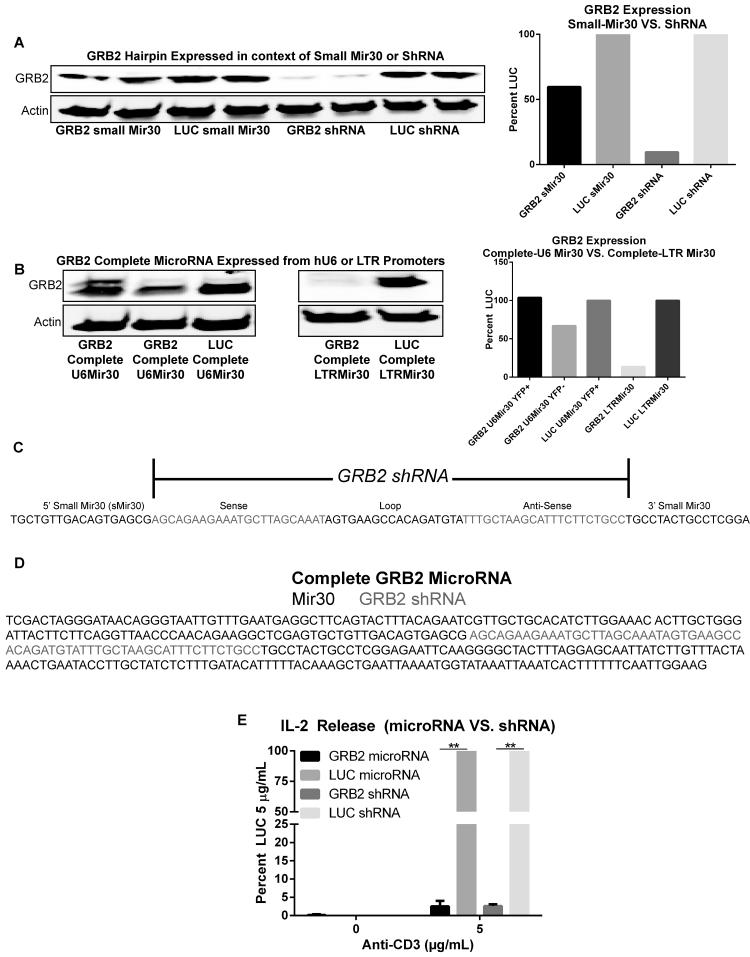
Lentiviruses allow efficient transduction of primary cells compared to LTR-based LMP retroviruses. However, in our hands the hU6 promoter utilized in pLK4A lentiviral constructs was only efficient if the small miR30-based hairpin extracted from the online algorithm was placed as a shRNA instead of microRNA (Figure 4a and c). Similar results were observed when HuT78 T cells were transduced with pLK4A lentiviruses expressing the complete (longer miR30 derived from LTR-LMP vectors) GRB2 microRNA constructs driven by the hU6 promoter (Figure 4b and d). Efficient suppression only occurred when the complete miR30 sequence was placed under the retroviral LTR promotor (Figure 4b and d). These results indicate that the suppressive efficacy of the microRNA is determined by the type of promoter that drives expression. Overall, effective suppression only occurred when the GRB2 shRNA alone was placed under the hU6 promoter or if the full microRNA was placed under the LTR promoter. However, utilizing the combination of the hU6-shRNA and LTR-complete-microRNA, we did not observe differences in T cell functions since our studies in HuT78 and primary CD4+ T cells was performed utilizing both LTR-miR30 and hU6-shRNA constructs had identical defects in cytokine release, and signal transduction16. Additionally HuT78 T cells lines transduced with hU6-GRB2shRNA or LTR-GRB2microRNA showed similar signaling defects including reduced IL-2 release, and enhanced phosphorylation of LCK, ZAP-70, and TCR ζ chain (Figure 4e, and data not shown).
Figure 4. hU6-induced GRB2 suppression is efficient only after removal of the full mir30 sequence.
(A) Left: HuT78 T cells transduced with pLKO.1 lentiviral constructs expressing hU6-driven GRB2 or LUC shRNAs flanked with or without small mir30 sequences. Right: Quantification of immunoblots. (B) Left: HuT78 T cells transduced with retro-LTR LMP or lentiviral-hU6 pLK4A constructs expressing GRB2 or LUC shRNAs flanked by the full mir30 sequence. Right: Quantification of immunoblots. Data represents 3 independent experiments with similar results. (C) Nucleotide sequence of GRB2 shRNA flanked by small mir30 sequence. (D) Nucleotide sequence of GRB2 shRNA flanked by the full mir30 sequence. (E) HuT78 T cells transduced with retroviruses carrying GRB2 or LUC microRNAs, and lentiviruses carrying hU6-GRB2 or LUC shRNAs were stimulated with 5 µg/mL anti-CD3 coated plates for 24 hrs. The supernatants were probed for IL-2 production via ELISA. The data was graphed as mean LUC secretions at 5 µg/mL anti-CD3 ±SEM from three independent experiments.
Transduced primary human CD4+ T cells are derived from a highly activated population
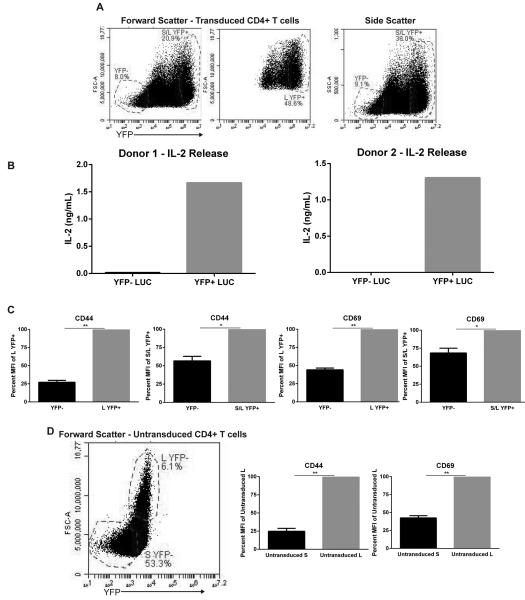
Upon analysis of transduced TCR-activated primary CD4+ T cells we observed differences in cellular size between the YFP− and YFP+ fractions. Specifically, the YFP+ fraction contained cells that were substantially larger in size as indicated by the forward and side scatter plots when compared to YFP− CD4+ T cells (Figure 5a). Based on these observations, we hypothesized that transduced YFP+ cells are more activated than cells with little to no detectable lentiviral integration. To demonstrate that lentiviruses infect highly-activated T cells more efficiently than cells not receiving efficient stimulatory signals, sorted YFP+ and YFP− CD4+ T cells were stimulated with 5 µg/mL plate-bound anti-CD3 for 24 hrs, and the levels of IL-2 were measured via ELISA. As expected, TCR-induced YFP+ cells were able to secrete substantially more IL-2 compared to YFP− T cells (Figure 5b). Additionally, expression of the activation markers CD44 and CD69 were substantially elevated in the YFP+ fraction, especially in the YFP+ fraction containing exclusively the larger cells (Figure 5c). This same large fraction also displayed higher activation markers in activated but untransduced cells (Figure 5d). Importantly, the latter indicates that the increase in size and activated phenotype was not a consequence of lentiviral integration. The increased YFP expression in larger transduced cells may be partially due to autofluorescence which stems from larger cell complexity. However, YFP autofluorescence in large untransduced cells is 2-4 orders of magnitude lower than fluorescence in large transduced cells (Figure 5a and d).
Figure 5. Transduced YFP+ are highly active and larger in size compared to YFP− primary CD4+ T cells.
(A) Activated primary CD4+ T cells were transduced with pLK4A lentiviruses carrying LUC shRNA and YFP for 3 days and then probed for YFP expression and forward/side scatter; S/L indicates Small/Large fractions. (B) Primary human CD4+ cells were transduced with YFP+ pLK4A lentiviruses expressing LUC shRNA. Sorted YFP+/- cells were stimulated with 5 µg/mL anti-CD3 coated plates for 24 hrs. The supernatants were probed for IL-2 production via ELISA. Illustrated is IL-2 production data from CD4+ primary cells obtained from two different donors. (C) Cells from “A” were surface stained with CD44 or CD69 conjugated with Alexa-fluor 647 and then gated on YFP−, S/L YFP+, and L YFP+ and then MFI was collected from 100,000 live gate events. Groups were graphed as ±SEM of YFP+ cells as indicated from three independent donors. (D) Untransduced activated primary CD4+ cells from the same donors as in “C” were stained with surface CD44 or CD69 conjugated with Alexa-fluor 647 and then gated on large “L” and small “S” fractions. Data was graphed as ±SEM of large cells from three independent donors.
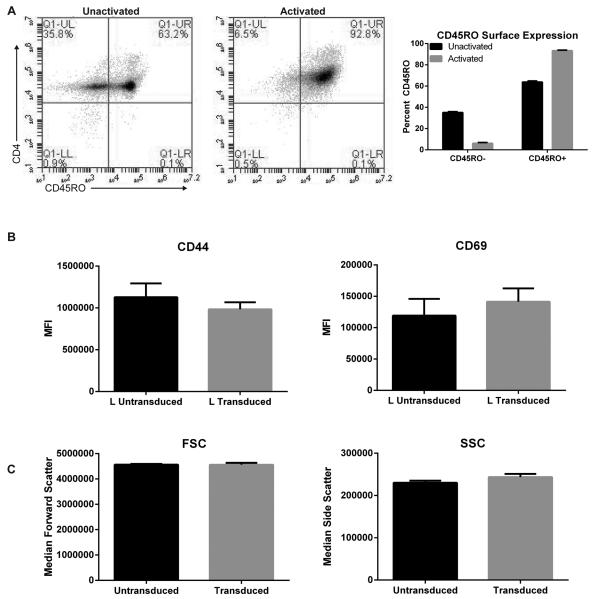
One explanation for the selective transduction of only a subset of CD4+ T cells is that not all cells respond to the CD3/CD28 signal because there is a polyclonal population of naïve and antigen experienced T cells. To test this idea, freshly isolated T cells and T cells activated for 6 days via CD3/CD28 were stained with CD45RO to differentiate between naïve (CD45RO negative) and antigen experienced (CD45RO positive) CD4+ T cells. Un-activated CD4+ T cells showed distinct CD45RO positive and CD45RO negative populations (Figure 6a). Upon activation via CD3/CD28, >90% of the cells expressed CD45RO, indicating that our method of stimulation activated the vast majority of T cells in culture (Figure 6a). Our results however suggest that most likely cells that were antigen-experienced prior to in vitro expansion were transduced better compared to cells that only received the in vitro signal. This is suggested by the two distinct populations stained with CD45RO prior to activation and transduced YFP+ cells in activated T cells (Figure 2a and 6a). Therefore, similar to antigen-inexperienced cells that were transduced, but with lower viral copy numbers as seen with reduced MFI (Figure 1b), we propose that the less activated “naïve” T cell population (Figure 6a) is not transduced as efficiently as the “primed” population. Importantly, in contrast to the enhanced activity of electroporated T cells, transduced YFP+ T cells did not show increased expression of T cell activation markers when compared to untransduced T cells (Figure 6b). Similarly, analysis of forward and side scatter did not show increased median cell size of transduced T cells when compared to untransduced counterparts (Figure 6c). Collectively, we demonstrate that in vitro TCR/CD28-induced stimulation of human primary CD4+ T cells efficiently activates T cells, however it does not produce a homogenous population, and only the highly activated T cells are efficiently transduced via lentiviruses.
Figure 6. Primary CD4+ T cells are adequately stimulated prior to transductions & untransduced/Transduced T cells show similar levels of T cell activation markers CD44/CD69.
(A) Left: Un-activated and 6 day activated CD4+ T cells were surface stained with CD4+ and CD45RO.1 and analyzed using flow cytometry. Right: Quantification of surface CD45RO in primary human CD4+ T cells pre and post activation from two independent donors. (B) Untransduced and transduced primary CD4+ T cells were stained with surface CD44 or CD69 conjugated with Alexa-fluor 647 and then gated on the Large “L” population. Data was graphed as mean MFI ±SEM from three independent donors. (C) Median forward “FSC” and side “SSC” scatters were quantified from untransduced and transduced primary CD4+ T cells. Data was graphed as mean Median FSC/SSC ±SEM from three independent donors.
DISCUSSION
In this study we tested multiple protocols to transduce human CD4+ T cells. We have described a method for the development of effective RNAi sequences that can produce near 100% protein suppression in human T cell lines and primary CD4+ T cells. Additionally, we highlighted a technique in which a mutated version of the suppressed protein can be added back simultaneously for further detailed analysis. MicroRNA/shRNAs can be generated quickly and integrated in lentiviral constructs within a week. Moreover, long-term use of this technology is more efficient and cost-effective when compared to the use of commercial siRNA or plasmid DNA transfections. One caveat of producing an in house microRNA/shRNAs is the need to empirically test each microRNA/shRNA to determine the suppression efficiency. The researcher must also be aware of off target effects stemming from long-term protein suppression in stable cell lines. The latter is true for both commercial and in house produced RNAi. To prevent potential misleading results from target effects, the researcher should first examine if the expression of proteins associated with and/or similar in sequence to the protein being targeted by the hairpins (i.e. proteins of the same family) are altered. Additionally, alternative microRNA/shRNAs that target a different sequence of the same protein, but would presumably have different off target effects, would provide confirmation of the results. We found that our method can generate multiple efficient hairpins targeting the same protein, even if it not as efficient as the primary hairpin (i.e. 60-80% protein suppression efficacy)16.
Upon development, recombinant DNA can then be delivered efficiently into activated and un-activated human CD4+ T cells utilizing lentiviral transductions. However, concentrated lentiviruses preps give the best transduction efficiency (percent integration) and number of gene copies (determined via YFP MFI) relative to unconcentrated preparations. Electroporation may also be used, however this method is highly toxic to cells, and should be used as an alternative method6, 8, 23. Additionally, stable cell lines produced via viral transductions are more convenient to use, and give reproducible results when compared to the need to repeatedly electroporate T cell lines with variable efficiencies between experiments. Viral-based production of stable cell lines also yield large amounts of cells carrying microRNA/shRNAs which is essential for assays requiring high cell numbers such as immunoprecipitation of proteins with low expression. Performing these types of experiments utilizing electroporated cells is not cost-effective and will require multiple electroporations for one replicate. Moreover, electroporation is not the optimal method for T cell analysis due to the increased activation phenotype of T cells induced by electroporation23. A recent study demonstrated that electroporated primary human CD4+ T cells had enhanced intracellular calcium levels, increased surface activation markers, augmented transcriptional activity, and amplified sensitivity to phytohemagglutinin (PHA)23. These observations indicate that assays examining T cell function (i.e. cytokine production, calcium mobilization, surface protein expression) may give misleading results when electroporation is used and are best performed in virally transduced T cells. In contrast, we found that lentiviral transduced primary CD4+ T cells express similar MFI levels of surface T cell activation markers CD44/CD69 and have similar cell morphology compared to untransduced cells (Figure 6b and c).
Next, there are currently new methods that could produce complete gene knockout in mammalian cells such as the RNA-based bacterial defense mechanism clustered regularly interspaced short palindromic repeats (CRISPR). This system utilizes a CAS9 endonuclease and a guide RNA (gRNA) molecule specific for invading bacteriophages and plasmid DNA that may integrate into the genome27, 28. In the case of mammalian cells, the CAS9-gRNA complex can be engineered to target specific dsDNA sequences in the genome. In contrast to RNAi technology that targets mRNA, effective CRISPR CAS9-gRNA can produce complete knockout of genes through frameshift mutations generated by insertion/deletions introduced by an error prone non-homologues end joining repair system27. Gene replacement may also occur through homology directed repair27, 29. Additionally, the CRISPR system has the advantage of simultaneously targeting multiple gene deletions in mice30.
Although knockout of genes utilizing CRISPR is an attractive approach, there are caveats associated with this method. First, the CRISPR system is inefficient at targeting genomic deletions in human CD4+ T cells. A recent study by Mandal et al demonstrated that a highly efficacious gRNA produced deletion of beta-2 microglobulin in up to 48% of HEK 293T cells, but only 5% of human CD4+ T cells31. Second, even after successful genomic deletion in human CD4+ T cells, off target effects may occur from deletion of DNA sequences similar to the sequence targeted by the gRNA. Recent studies in human cells demonstrated off target deletions occurring in the genome, even with sites that differ by five nucleotides from the target DNA32, 33, 34. Similarly, gRNA with a one base-pair mismatch to the target DNA allows CAS9 to cleave the target DNA35, 36. Therefore, utilizing CRISPR requires screening the genome for similar sites to the gRNA prior to and after deletion of the target gene. These effects can produce misleading results especially in genes expressing homologous proteins such as the GRB2 family of adaptors or with DNA that produces splice variants with different functions (i.e. GRB2 and GRB3-3)37. In the end, the use of CRISPR system would require the screening of numerous clones of human T cells, which would be feasible for cell lines but not short lived primary human T cells. Therefore, given the above caveats, studies in primary human CD4+ T cells should be performed utilizing established methods such as the microRNA/shRNA based methods presented in this study.
We have also demonstrated that in contrast to the LTR, the hU6 promoter can be inefficient if the RNAi used was expressed in the context of microRNA. The reasons for this discrepancy are not completely clear, but could be a result of one or a combination of the following. First, in LMP retroviruses, the distance from the LTR promotor to the microRNA (>100 bp) is much greater than the distance between the lentiviral hU6 promoter and its transcripts (11 bp). The increased space found after the LTR promoter could lead to higher microRNA hairpin stability in the course of initiation and formation of the hairpin that results in efficient processing by the RNAi machinery. Second, the formation of the microRNA hairpin from the hU6 promoter may not be efficient due to intrinsic mechanistic differences between the Type II and III polymerases utilized by LTR and hU6 promoters, respectively. The cell type in which the promoter is expressed may also account for some of these differences38, 39, 40, 41, 42. Discrepancies in gene silencing between distinct promoters were also recently demonstrated by Roelz et al. The authors demonstrate that shRNA expression from the hU6 promoter displayed a fourfold increase in inhibition efficacy when compared to the same shRNA driven by the murine U6 promoter in both human and murine cells40. Therefore, reduction of transcripts and/or inefficiency in hairpin formation may play a role in determining the final inhibition effects. Overall, we describe clear functional differences between LTR/hU6 microRNA/shRNA-induced protein suppression. Our data demonstrate that caution must be taken in the development and interchange of microRNA/shRNAs between multiple promoters. We, therefore, suggest that the safest method is to first screen for shRNAs and then embed efficient sequences in the context of microRNAs.
Interestingly, although our method of stimulation activates the vast majority of T cells as determined by CD45RO expression, there appear to be distinct populations with different activation phenotypes. Specifically, cells that were highly activated were selectively transduced with high efficiency leading to greater gene expression from the viral vectors. These highly transduced cells had increased cytokine secretion, and CD44/CD69 upregulation. Our results highlight an important point, that in vitro activated T cells are heterogeneous and do not receive similar strength of stimulatory signals in culture. This may produce misleading results, especially if stimulated populations of a transduced culture were to be compared for activation status based on protein inhibition effects and/or protein expression. In this case, cells that had incorporated larger numbers of viral genomes will be intrinsically activated prior to transductions and subsequently the data will provide misleading results. This should be taken into consideration during experimental planning and analysis. Overall, we present optimized protocols that should significantly enhance the quality of DNA integration into primary human T cells. This is important as studies in murine T cells or human cell lines do not always produce similar results when compared to the more physiological primary human CD4+ T cells4, 5, 43, 44.
MATERIALS AND METHODS
Purification and growth of human CD4+ peripheral blood T cells
Peripheral blood mononuclear cells (PBMC) were obtained from whole blood of healthy donors using leukocyte reduction system (LRS) cones as previously described45_ENREF_20. Blood donors have consented for blood donation at the DeGowin Blood Center at the University of Iowa Hospitals and Clinics. The consent allows peripheral blood cells not used for transfusion to be used for research at the University of Iowa. The consent process and documents for these donors were approved by the IRB for the University of Iowa. Because all cells used in these studies were obtained from normally discarded products, the donors approved for the use of their cells in research projects and the donors were completely de-identified, these studies were exempt from further IRB approval.
Negative selection of primary human CD4+ T cells was performed using CD4+ T cell isolation kit II (Miltenyi Biotec) resulting in >98% purity. Isolated CD4+ T cells were subsequently activated with magnetic Dyna beads (Invitrogen) crosslinked with anti-CD3 (OKT3, BioLegend) and anti-CD28 (CD28.2, BioLegend) in the presence of 100 U/mL recombinant IL-2. Cells were cultured at 37°C and 5% CO2 in complete RPMI 1640 (RPMI medium supplemented with 10% FBS, 50 U/mL penicillin, 50 μg/mL streptomycin, and 2mM l-glutamine) (Gibco). Prior to cytokine release assays, cells were rested for 1 day in complete RPMI without magnetic beads or IL-2. For flow cytometry analysis or cell sorting, cells were removed from the magnetic beads and immediately analyzed or sorted.
Growth of HuT78 and E6.1 T cell lines
HuT78 CD4+ T cells were acquired directly from ATCC (TIB-161). HuT78 CD4+ T cell lines were cultured at 37°C and 5% CO2 in complete IMDM medium (IMDM medium supplemented with 20% FBS, 50 U/mL penicillin, 50 μg/mL streptomycin, and 2mM l-glutamine) (Gibco). Retro/lentiviral transduced cell lines were kept in selection in the presence of 1-2 µg/mL puromycin. Jurkat E6.1 T cells were cultured in complete RPMI medium. For immunoblot analysis, the cells were lysed with the addition of 4-fold excess of hot 2X lysis buffer (20 mM Tris pH8.0, 2 mM EDTA, 2 mM Na3VO4, 20 mM DTT, 2% SDS, and 20% glycerol). Lysates were then heated to 95°C for 4 min and sonicated to reduce viscosity.
Cytokine detection
Primary CD4+ T cells were washed in complete RPMI 1640, and then resuspended at 2 × 105 cells/mL. Cells were stimulated by adding 0.5 mL of cell suspension to 24 well plates coated with 5 µg/mL of anti-CD3 for 24 hrs. IL-2 protein concentrations in the culture supernatants were measured using standard TMB ELISA utilizing a spectrophotometric plate reader with a reading absorbance at 450 nm.
Immunoblotting
Cellular lysates were loaded onto a 4-15% precast Criterion polyacrylamide gel (Biorad). The separated proteins were transferred onto PVDF membranes (Millipore), blocked for 1 hr at room temperature with PBS-SEA Block buffer (Thermo Scientific). The PVDF membranes were then incubated with primary antibodies against GFP(YFP) (clone B-2, Santa Cruz Biotechnology, GRB2 (clone 23, Santa Cruz Biotechnology), LCK (Cell Signaling), Actin (clone C4, Millipore), or GAPDH (Meridian Life Sciences). Secondary antibodies conjugated to IRDye 800CW or IRDye 680 were diluted in SEA Block and incubated with the PVDF membranes for 30 min at room temperature. The membranes were then washed with PBS-0.1%SDS solution and then visualized using the Licor Odyssey Infrared detector.
Flow cytometry
2 × 106 cells were washed in FACS buffer (PBS, 10% FBS, and 0.05% sodium azide), and then resuspended in FACS buffer to a concentration of 1 × 106 cells/mL to probe GFP/YFP expression. For surface staining, primary CD4+ T cells were washed in FACs buffer, and then stained with anti-CD44 or anti-CD69 conjugated to Alexa-fluor 647 (Biolegend), CD45RO conjugated to PE-Cy5 (BD Pharmingen). For CD4 staining, cells were first stained with primary anti-CD4 (clone RPA-T4, Biolegend) and then with secondary Alexa 488 (Biolegend). Cells were left on ice for 30 min during staining while gently vortexing every 10 minutes. Cells were washed, and the mean fluorescence intensity (MFI) of each sample was obtained using Accuri C6 flow cytometer.
Quantification of data produced via flow cytometry and ELISA
IL-2 release data from HuT78 T cells was normalized to percent of maximum (max = 100%) control RNAi (LUC) stimulation. For flow cytometry, the data was normalized to percent of maximum controls as indicated on the Y-axis. After the MFI or IL-2 values were normalized for each independent experiment, multiple experiments were compiled together for a final average and SEM values. The following formula was utilized for normalization and quantification of the MFI values, and IL-2 release data for each data point:
-Data point as Percent of maximum of LUC stimulation (or as indicated on Y-axis) = (Raw value − Raw unstimulated value of control LUC or as indicated on Y-axis) ÷ (Raw maximum value of control LUC − Raw unstimulated value of control LUC or as indicated on Y-axis)
Vectors
Lentiviral pLK4 vectors were a kind gift from Dr. Stephen Bunnell._ENREF_3 pLKO.1 lentiviral vectors were obtained from Addgene (plasmid 8453)46. MSCV-LTRmiR30-PIG (LMP) retroviral expression vector and VSV-G were obtained from Dr. Bruce Hostager. Packaging plasmids pCL-Eco and Pax2 were donated by Drs. John Colgan and Dawn Quelle, respectively.
Cloning of RNAi into LMP and pLK4A retroviral vectors
Retroviral LMP vectors contain XhoI and EcoRI cloning site allowing the insertion of shRNAs in the context of endogenous mir30 driven by an LTR-based promoter. However, because microRNA-carrying LMP vectors are not efficient at infecting primary human CD4+ T cells (data not shown), shRNA-carrying pLK4A vectors were utilized for the transduction of these cells. In addition, the pLK4A vector allows for cloning of shRNA and add-back of wild-type and mutant proteins in T cells. Although microRNAs mimic physiological silencing mechanisms and should be the first choice, shRNAs in lentiviruses were utilized due to the inefficiency of the hU6 promoter to drive microRNAs (see Differential efficiency of LTR and hU6 in RNAi-mediated protein suppression). To accomplish this, shRNA sequences were first cloned into pLKO.1 vectors using AgeI and EcoRI sites followed by terminal thymidines that stop the transcriptional activity of the RNA polymerase.
MicroRNA/shRNA sequences were generated by using the online-based http://katahdin.cshl.edu (or other online RNAi generating algorithms). Upon entering the online program, click on si/shRNA retriever, choose number of oligos, and species type. Next, generate microRNAs by either entering the accession (NM_mRNA) or the full sequence of target mRNA, and then click Retrieve Oligo. Listed below are microRNAs generated using the above algorithm. Note that in this case the flanking mir-30 sequences generated by the algorithm were removed and the shRNA portion is illustrated (Sense, Loop, Anti-Sense).
GRB2: AGCAGAAGAAATGCTTAGCAAATAGTGAAGCCACAGATGTATTTGCTAAGCATTTCTTCTGCC
LCK: ACCCATCTACATCATCACTGAATAGTGAAGCCACAGATGTATTCAGTGATGATGTAGATGGGC
The oligonucleotides were then separated into forward and reverse sequences capable of annealing with 10 bp in the loop sequence (at the center of the microRNA/shRNA). The following primers were utilized to create the complete GRB2 hairpin:
Complete GRB2 shRNA with flanking AgeI/EcoRI and Terminal thymidines X6: GAACCTACCGGTAGCAGAAGAAATGCTTAGCAAATAGTGAAGCCACAGATGTATTTGCTAAGCATTTCTTCTGCCTTTTTTGAATTCGGAAC
GRB2 F primer for Klenow reaction: GAACCTACCGGTAGCAGAAGAAATGCTTAGCAAATAGTGAAGCCACAG
GRB2 R primer for Klenow reaction: GTTCCGAATTCAAAAAAGGCAGAAGAAATGCTTAGCAAATACATCTGTGGCTTC
The F/R sequences were extended into the complete hairpin with a Klenow cycle. The reaction was incubated for at 30°C 30 min to generate double stranded oligos, and then at 75°C for 20 min to inactivate Klenow. The shRNAs were ligated into pLKO.1 vectors, transformed, and sequenced to determine positive clones.
Next, to move the U6-shRNA cassette from pLKO.1 into the final primary CD4+ T cell transduction vector, pLK4A, sequences from pLKO.1 were amplified and cloned into SpeI and PvuI site in the pLK4A lentiviral vectors using the following primers:
GRB2 pLK4A-SpeI F: CAACCAACTAGTGAGGGCCTATTTCCCATGATTCCTTCATATTTGC
GRB2 pLK4A-PvuI R: TGGTTGCGATCGAAAAAAGGCAGAAGAAATGCTTAGCAAATACATCTG
To generate mutated proteins (GRB2 or GADS) that are resistant to shRNA mediated inhibition, human GRB2 and GADS cDNA was mutated using primers containing shRNA-sense bases to produce GRB2 or GADS cDNA with a different nucleotide sequence that produces an identical amino acid sequence to the wild-type protein. Both the GRB2 and GADS variants were amplified with AgeI and NotI primers, and then ligated into pLK4A vectors containing GRB2 or GADS shRNAs.
Viral production and purification
18-24 hrs prior to transfections, 3.5 × 106 or 7.5 × 106 293T cells were seeded in 10 or 15 cm culture dishes at 37°C and 5% CO2 in complete DMEM medium (DMEM medium supplemented with 10% FBS, 50 U/mL penicillin, 50 μg/mL streptomycin, 1X MEM NEAA, and 2mM l-glutamine) (Gibco). On the day of transfection the medium was replaced with 5 or 16 mL of fresh complete DMEM.
To generate retro or lentiviral particles, 293T cells were transfected utilizing calcium phosphate method. In 10 cm plates (For 15 cm plates multiply reagent value by 2), plasmids (LMP/pLK4A 15 μg, pCL-Eco/Pax2 10 μg, VSV-G 7.5 μg) were mixed in 0.5 mL of 1X HBS buffer (Hepes free acid 21 mM, NaCl 137 mM, D+ Glucose monohydrate 5 mM, KCl 50 mM, Na2HPO4 0.35 mM, pH 7.5). Next, 30 μL of sterile 2.5 M CaCl2 was added drop-wise to the HBS-DNA solution, mixed by pipetting, and incubated for 20-30 min at room temperature. The HBS-DNA-CaCl2 mix was then added drop-wise around the 293T plate and then incubated for 15-20 hrs.
Next, transfection medium was replaced with 10 or 20 mL of fresh medium for 10 or 15 cm plates, respectively. Because some viral production initiates shortly after transfection, we recommend that the transfection medium be replaced at earlier time points to prevent viral loss. Viral containing supernatant was subsequently harvested every 24 hrs for 2 days, and filtered through 0.45 μm Durapore Millex (Millipore) filters. Filtered supernatant was then divided into round-bottom polycarbonate high-speed tubes (Nalgene-Oak ridge). The tubes were centrifuged at 4°C for 1.5-2 hrs at 48,000 × g in Sorvall RC6-Plus centrifuge (SS-34 Rotor). The tubes were handled carefully as to not resuspend the viral pellet, and the supernatant was aspirated using sterile glass Pasteur pipettes. The pellets were resuspended in 0.5-2mL complete RPMI.
Transduction and development of stable HuT78 T cell lines
Human HuT78 CD4+ T cells have similar TCR signaling kinetics to primary human CD4+ T cells when compared to other cell lines47. For this reason, we optimized transductions of this cell line to utilize HuT78 T cells as a model for understanding T cell signaling. For HuT78 T cell transductions, 4-5 × 105 cells were incubated upright in 1-1.5 mL of concentrated viral supernatant in 25 cm2 flasks in the presence of 8 μg/mL Hexadimethrine bromide (Sigma Aldrich) with periodical mixing. Upright incubation allows the small volume of viral medium to spread on a small surface during transduction. Based on our observations, we speculate that this method allows higher probability of virus:cell interaction and subsequent increased frequency of transduction. The viral quantity is equivalent to medium obtained from 1-2 10 cm culture dishes. Generally, transductions are allowed to continue until the phenol red indicator in the tissue culture medium becomes visibly yellow (indicating proliferation of cells). This takes place in a period of 48-72 hrs depending on the number of cells used. Post transduction, HuT78 T cells were resuspended in fresh complete IMDM with 0.5 μg/mL of puromycin and cells were allowed to expand. The expansion is determined by the visible phenol red color changes of the complete IMDM medium (yellow) which usually takes 2-4 days depending on the number of cells used and efficiency of transduction. Puromycin is then increased gradually (0.5 μg/mL increments) to 1.5-2 μg/mL after each cell passage before examining protein inhibition and/or levels of protein expression. Similar to utilizing viral concentrates, duration of transductions may significantly impact the efficiency of viral integration. Therefore, the duration of virus:cell interaction should in general be lengthened rather than shortened. We found that methods such as spinfection are unnecessary for the enhancement of gene delivery to human T cells (data not shown). Rather it is the concentration of viruses and duration of infection that carry a major impact on efficiency of gene delivery.
Transduction of primary human CD4+ T cells
For primary human CD4+ T cell transductions, purified cells were activated for 1 day with magnetic Dyna beads containing anti-CD3 and anti-CD28 in the presence of IL-2. We have found that the duration of activation does not significantly reduce transduction efficiency. However, it is better to transduce cells during initial activation prior to expansion (12-24 hrs post activation) to enrich transduced cells that are proliferating in culture. Next, 10-25 × 106 cells were incubated upright in 1-1.5 mL of concentrated viral supernatant in 25 cm2 flasks in the presence of 8 μg/mL Hexadimethrine bromide (Sigma Aldrich) with periodical mixing. The viral quantity for the transduction of 25 × 106 cells is equivalent to medium obtained from 8-12 10 cm or 4-6 15 cm culture dishes. 100 U/mL of IL-2 plus beads containing anti-CD3 and anti-CD28 were present during the transduction to preserve the activation status of the cells. 2-5 mL of complete RPMI medium, Hexadimethrine bromide, and IL-2 were added to the cells every 24 hrs or after medium becomes visibly yellow. At this point the flasks were placed horizontally to reduce cell packing and allow better expansion of the cells. After 72-96 hrs cells were resuspended in fresh complete RPMI without the beads or IL-2 for analysis.
Transduction of antigen inexperienced T cells
Primary CD4+ T cells were cultured in the presence of 200 U/mL IL-2 for 12-24 hrs. 5-10 × 106 cells were then transduced by incubating upright with 1-1.5 mL concentrated lentiviral supernatant in the presence of 8 μg/mL Hexadimethrine bromide. The viral quantity for the transduction of 10 × 106 cells is equivalent to medium obtained from 4-6 10 cm or 2-3 15 cm culture dishes. Higher concentration of lentiviruses correlates with increased transduction efficiency (data not shown). The transductions occurred over a period of 3-6 days with periodical mixing and without adding extra medium. In contrast to activated cells, after the first day of transduction, transduced antigen inexperienced T cells should not be placed horizontally as they are not in state of expansion. This allows the cells to have a better interaction with lentiviruses. After transduction was complete, the viral supernatant was removed and replaced with fresh medium containing IL-2, and YFP or protein expression was assessed via flow cytometry and/or immunoblot.
Electroporation of primary human CD4+ T cells
Primary human CD4+ T cells were activated for 1-4 days and then magnetic beads were removed prior to transfections. 5 × 106 CD4+ T cell were placed in 100 µL electroporation buffer using buffers optimized in Chicaybam et al (1M, 1SM, or 3P)6. Experimentation with these reagents is recommended to determine buffers which produce minimal toxicity and highest transfection efficiency. Next, 5-20 µg of a plasmid carrying GFP was added to the cell reagent solution. If using a plasmid that does not carry a fluorescent marker, we recommend co-transfection with a GFP carrying plasmid in order to determine the general transfection efficiency. Immediately, the cell-reagent mix was aliquoted into a 0.2 cm Lonza or Bio-Rad Gene Pulser cuvette. The cells were electroporated using Lonza’s T cell programs (such as T-23 or U-14). Experimentation with these programs is also recommended to determine which produce least cell death, and highest transfection efficiency. Cells were immediately removed from the cuvette and then cultured in 5 mL complete RPMI medium in the presence of 100 U/mL IL-2. GFP fluorescence was analyzed after 24-48 hrs post transfection.
Statistical analysis
Analysis of flow cytometry surface staining and ELISA were performed in GraphPad prism software using two-tailed t-test assuming unequal variance. Levels of significance p<.05 and p<.01 are presented as * and **, respectively.
Supplementary Material
Acknowledgment
Some of the data presented herein were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine Core Research Facilities/ Holden Comprehensive Cancer Center Core Laboratory at the University of Iowa. The Facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran's Administration Medical Center. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Human rIL-2 from Dr. Maurice Gately, Hoffmann - La Roche Inc. This work was supported by National Institutes of Health Predoctoral Training Grant in Immunology (5T32 AI007485). These studies were supported by R01 CA136729 and R01 CA136729-S1 from the National Institutes of Health to JCDH.
REFERENCES
- 1.Basson MA, Zamoyska R. Insights into T-cell development from studies using transgenic and knockout mice. Molecular biotechnology. 2001;18(1):11–23. doi: 10.1385/MB:18:1:11. [DOI] [PubMed] [Google Scholar]
- 2.Mak TW, Penninger JM, Ohashi PS. Knockout mice: a paradigm shift in modern immunology. Nature reviews Immunology. 2001;1(1):11–19. doi: 10.1038/3509551. [DOI] [PubMed] [Google Scholar]
- 3.Jang IK, Zhang J, Chiang YJ, Kole HK, Cronshaw DG, Zou Y, et al. Grb2 functions at the top of the T-cell antigen receptor-induced tyrosine kinase cascade to control thymic selection. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(23):10620–10625. doi: 10.1073/pnas.0905039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. Journal of immunology. 2004;172(5):2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 5.Shay T, Jojic V, Zuk O, Rothamel K, Puyraimond-Zemmour D, Feng T, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(8):2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chicaybam L, Sodre AL, Curzio BA, Bonamino MH. An efficient low cost method for gene transfer to T lymphocytes. PloS one. 2013;8(3):e60298. doi: 10.1371/journal.pone.0060298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TK, Eberwine JH. Mammalian cell transfection: the present and the future. Analytical and bioanalytical chemistry. 2010;397(8):3173–3178. doi: 10.1007/s00216-010-3821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebert O, Finke S, Salahi A, Herrmann M, Trojaneck B, Lefterova P, et al. Lymphocyte apoptosis: induction by gene transfer techniques. Gene therapy. 1997;4(4):296–302. doi: 10.1038/sj.gt.3300394. [DOI] [PubMed] [Google Scholar]
- 9.Costello E, Munoz M, Buetti E, Meylan PR, Diggelmann H, Thali M. Gene transfer into stimulated and unstimulated T lymphocytes by HIV-1-derived lentiviral vectors. Gene therapy. 2000;7(7):596–604. doi: 10.1038/sj.gt.3301135. [DOI] [PubMed] [Google Scholar]
- 10.Cavalieri S, Cazzaniga S, Geuna M, Magnani Z, Bordignon C, Naldini L, et al. Human T lymphocytes transduced by lentiviral vectors in the absence of TCR activation maintain an intact immune competence. Blood. 2003;102(2):497–505. doi: 10.1182/blood-2003-01-0297. [DOI] [PubMed] [Google Scholar]
- 11.Cooray S, Howe SJ, Thrasher AJ. Retrovirus and lentivirus vector design and methods of cell conditioning. Methods in enzymology. 2012;507:29–57. doi: 10.1016/B978-0-12-386509-0.00003-X. [DOI] [PubMed] [Google Scholar]
- 12.Charrier S, Dupre L, Scaramuzza S, Jeanson-Leh L, Blundell MP, Danos O, et al. Lentiviral vectors targeting WASp expression to hematopoietic cells, efficiently transduce and correct cells from WAS patients. Gene therapy. 2007;14(5):415–428. doi: 10.1038/sj.gt.3302863. [DOI] [PubMed] [Google Scholar]
- 13.Brenner S, Malech HL. Current developments in the design of onco-retrovirus and lentivirus vector systems for hematopoietic cell gene therapy. Biochimica et biophysica acta. 2003;1640(1):1–24. doi: 10.1016/s0167-4889(03)00024-7. [DOI] [PubMed] [Google Scholar]
- 14.Ramezani A, Hawley TS, Hawley RG. Stable gammaretroviral vector expression during embryonic stem cell-derived in vitro hematopoietic development. Molecular therapy : the journal of the American Society of Gene Therapy. 2006;14(2):245–254. doi: 10.1016/j.ymthe.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilal MY, Zhang EY, Dinkel B, Hardy D, Yankee TM, Houtman JC. GADS is required for TCR-mediated calcium influx and cytokine release, but not cellular adhesion, in human T cells. Cellular signalling. 2015;27(4):841–850. doi: 10.1016/j.cellsig.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bilal MY, Houtman JC. GRB2 Nucleates T Cell Receptor-Mediated LAT Clusters That Control PLC-gamma1 Activation and Cytokine Production. Frontiers in immunology. 2015;6:141. doi: 10.3389/fimmu.2015.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kesireddy V, van der Ven PF, Furst DO. Multipurpose modular lentiviral vectors for RNA interference and transgene expression. Molecular biology reports. 2010;37(6):2863–2870. doi: 10.1007/s11033-009-9840-8. [DOI] [PubMed] [Google Scholar]
- 18.Keck K, Volper EM, Spengler RM, Long DD, Chan CY, Ding Y, et al. Rational design leads to more potent RNA interference against hepatitis B virus: factors effecting silencing efficiency. Molecular therapy : the journal of the American Society of Gene Therapy. 2009;17(3):538–547. doi: 10.1038/mt.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nature biotechnology. 2004;22(3):326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 20.Davidson BL, McCray PB., Jr. Current prospects for RNA interference-based therapies. Nature reviews Genetics. 2011;12(5):329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic acids research. 2004;32(3):1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M, Ma Z, Selliah N, Weiss G, Genin A, Finkel TH, et al. The impact of Nucleofection(R) on the activation state of primary human CD4 T cells. Journal of immunological methods. 2014;408:123–131. doi: 10.1016/j.jim.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huls MH, Figliola MJ, Dawson MJ, Olivares S, Kebriaei P, Shpall EJ, et al. Clinical application of Sleeping Beauty and artificial antigen presenting cells to genetically modify T cells from peripheral and umbilical cord blood. Journal of visualized experiments : JoVE. 2013;(72):e50070. doi: 10.3791/50070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis BL, Potts PR, Porteus MH. Creating higher titer lentivirus with caffeine. Human gene therapy. 2011;22(1):93–100. doi: 10.1089/hum.2010.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen JC, Sechelski J. Use of sodium butyrate to enhance production of retroviral vectors expressing CFTR cDNA. Human gene therapy. 1995;6(9):1195–1202. doi: 10.1089/hum.1995.6.9-1195. [DOI] [PubMed] [Google Scholar]
- 27.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 28.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annual review of genetics. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 29.Zheng Q, Cai X, Tan MH, Schaffert S, Arnold CP, Gong X, et al. Precise gene deletion and replacement using the CRISPR/Cas9 system in human cells. BioTechniques. 2014;57(3):115–124. doi: 10.2144/000114196. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandal PK, Ferreira LM, Collins R, Meissner TB, Boutwell CL, Friesen M, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell stem cell. 2014;15(5):643–652. doi: 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature biotechnology. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature biotechnology. 2013;31(9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romero F, Ramos-Morales F, Dominguez A, Rios RM, Schweighoffer F, Tocque B, et al. Grb2 and its apoptotic isoform Grb3-3 associate with heterogeneous nuclear ribonucleoprotein C, and these interactions are modulated by poly(U) RNA. The Journal of biological chemistry. 1998;273(13):7776–7781. doi: 10.1074/jbc.273.13.7776. [DOI] [PubMed] [Google Scholar]
- 38.Das G, Henning D, Reddy R. Structure, organization, and transcription of Drosophila U6 small nuclear RNA genes. The Journal of biological chemistry. 1987;262(3):1187–1193. [PubMed] [Google Scholar]
- 39.Wise TG, Schafer DJ, Lambeth LS, Tyack SG, Bruce MP, Moore RJ, et al. Characterization and comparison of chicken U6 promoters for the expression of short hairpin RNAs. Animal biotechnology. 2007;18(3):153–162. doi: 10.1080/10495390600867515. [DOI] [PubMed] [Google Scholar]
- 40.Roelz R, Pilz IH, Mutschler M, Pahl HL. Of mice and men: human RNA polymerase III promoter U6 is more efficient than its murine homologue for shRNA expression from a lentiviral vector in both human and murine progenitor cells. Experimental hematology. 2010;38(9):792–797. doi: 10.1016/j.exphem.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Makinen PI, Koponen JK, Karkkainen AM, Malm TM, Pulkkinen KH, Koistinaho J, et al. Stable RNA interference: comparison of U6 and H1 promoters in endothelial cells and in mouse brain. The journal of gene medicine. 2006;8(4):433–441. doi: 10.1002/jgm.860. [DOI] [PubMed] [Google Scholar]
- 42.Lebbink RJ, Lowe M, Chan T, Khine H, Wang X, McManus MT. Polymerase II promoter strength determines efficacy of microRNA adapted shRNAs. PloS one. 2011;6(10):e26213. doi: 10.1371/journal.pone.0026213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Molecular & cellular proteomics : MCP. 2009;8(3):443–450. doi: 10.1074/mcp.M800258-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur G, Dufour JM. Cell lines: Valuable tools or useless artifacts. Spermatogenesis. 2012;2(1):1–5. doi: 10.4161/spmg.19885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tremblay MM, Houtman JC. TCR-mediated functions are enhanced in activated peripheral blood T cells isolated from leucocyte reduction systems. Journal of immunological methods. 2014 doi: 10.1016/j.jim.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. Rna. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartelt RR, Cruz-Orcutt N, Collins M, Houtman JC. Comparison of T cell receptor-induced proximal signaling and downstream functions in immortalized and primary T cells. PloS one. 2009;4(5):e5430. doi: 10.1371/journal.pone.0005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.